Concepts
Overview of how data is organized!
If you want help development, read carefully the OpenDataBio data model concept before start collaborating.
To facilitate the understanding of the concepts, as include many tables and complex relationships, the OpenDataBio data model is divided in the four categories listed below.
1 - Core Objects
Objects that may have Measurements from custom Traits!
Core objects are: Location, Voucher, Individual and Taxon. These entities are considered “Core” because they may have Measurements, i.e. you may register values for any custom Trait.
The Individual object refer to Individual organism that have been observed once (an occurrence) or has been tagged for monitoring, such as tree in a permanent plot, a banded bird, a radio-tracked bat. Individuals may have one or more Vouchersin a BioCollection, and one or multiple Locations, and will have a taxonomic Identification. Any attribute measured or taken for individual organism may be associated with this object through the Measurement Model model.
The Voucherobject is for records of specimens from Individuals deposited in a Biological Collection. The taxonomic Identification and the Location of a Voucher is that of the Individual it belongs to. Measurements may be linked to a Voucher when you want to explicitly register the data to that particular sample (e.g. morphological measurements; a molecular marker from an extraction of a sample in a tissue collection). Otherwise you could just record the Measurement for the Individual the Voucher belongs to. The voucher model is also available as special type of Trait, the LinkType, making it possible to record counts for the voucher’s Taxon at a particular Location.
The Location object contains spatial geometries, like points and polygons, and include plots and transects as special cases. An Individual may have one location (e.g. a plant) or more locations (e.g. a monitored animal). Plots and Transect locations may be registered as a spatial geometry or only point geometry, and may have Cartesian dimensions (meters) registered. Individuals may also have Cartesian positions (X and Y or Angle and Distance) relative to their Location, allowing to account for traditional mapping of individuals in sampling units. Ecological relevant measurements, such as soil or climate data are examples of measurements that may be linked to locations Measurement.
The Taxon object in addition to its use for the Identification of Individuals, may receive Measurements, allowing the organization of secondary, published data, or any kind of information linked to a Taxonomic name. A BibReference may be included to indicate the data source. Moreover, the Taxon model is available as special type of Trait, the LinkType, making it possible to record counts for Taxons at a particular Location.
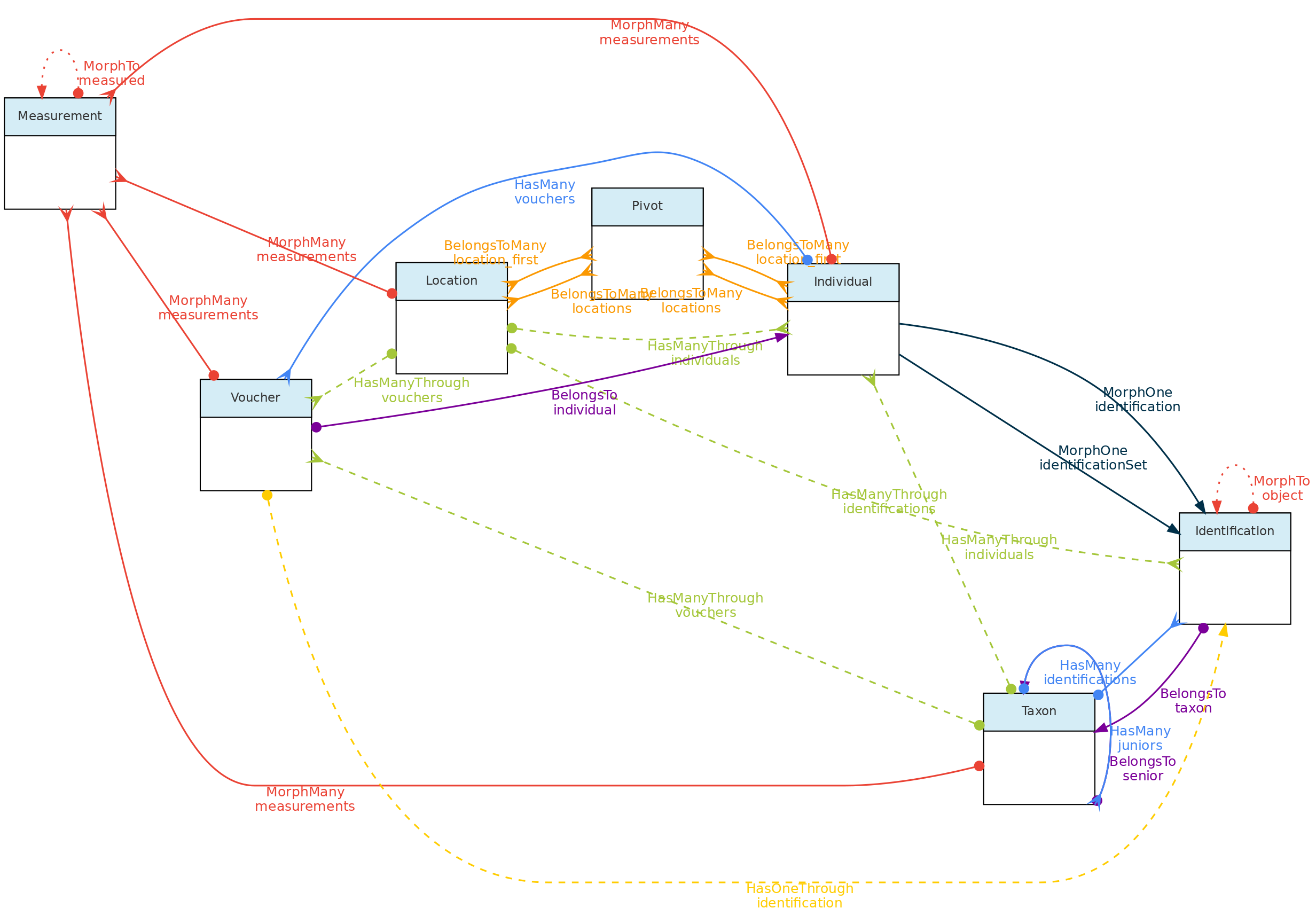
This figure show the relationships among the Core objects and with the Measurement Model. The Identification Model is also included for clarity. Solid links are direct relationships, while dashed links are indirect relationships (e.g. Taxons has many Vouchers through Individuals, and have many Individuals through identifications). The red solid lines link the Core objects with the Measurement model through polymorphic relationships. The dotted lines on the Measurement model just allow access to the measured core-object and to the models of link type traits.
Location Model
The Locations table stores data representing real world locations. They may be countries, cities, conservation units, or any spatial polygon, point or linestring on the surface of Earth. These objects are hierarchical and have a parent-child relationship implemented using the Nested Set Model for hierarchical data of the Laravel library Baum and facilite both validation and queries.
Special location types are plots and transects, which together with point locations allow different sampling methods used in biodiversity studies. These Location types may also be linked a parent location and in addition also to three additional types of location that may span different administrative boundaries, such as Conservation Units, Indigenous Territories and any Environmental layer representing vegetation classes, soil classes, etc…with defined spatial geometries.
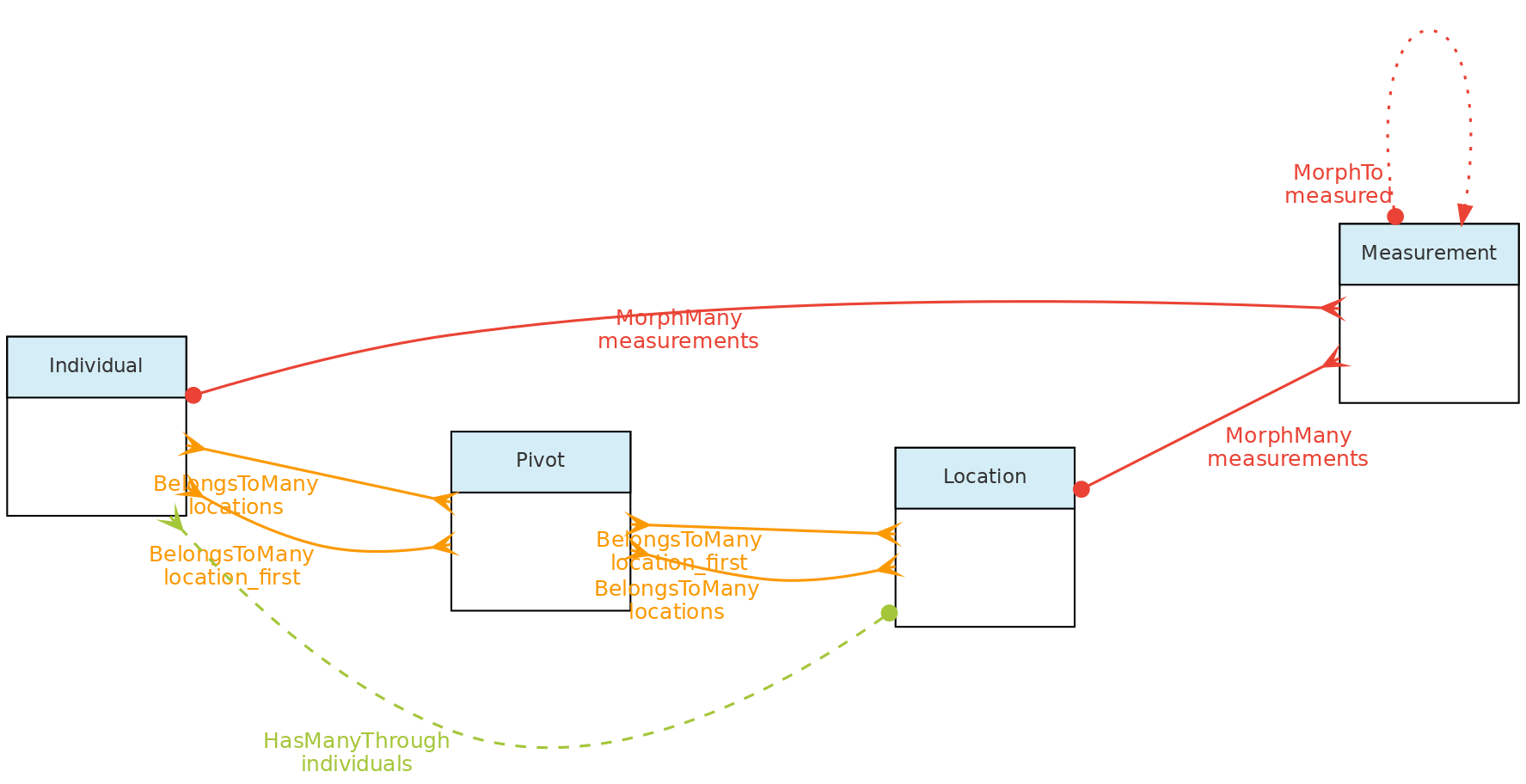 This figure shows the relationships of the
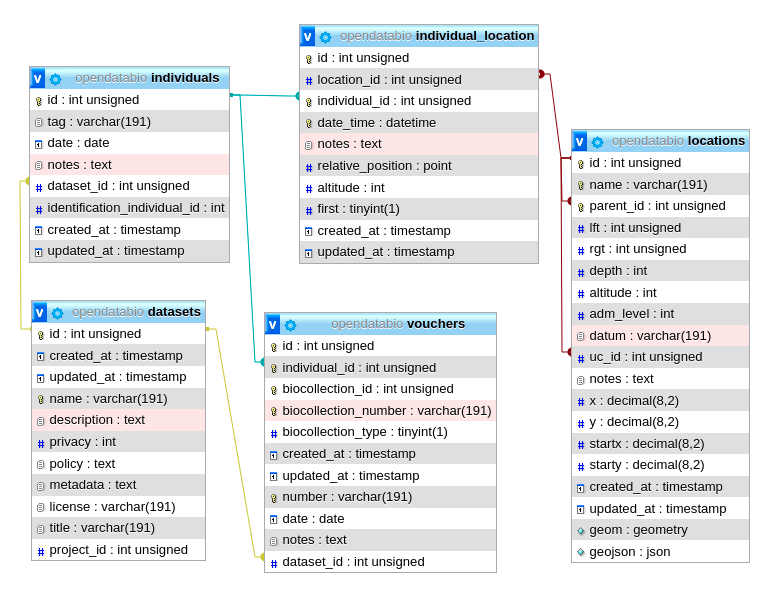
This figure shows the relationships of the Location model throught the methods implemented in the shown classes. The pivot table linking Location to Individual allow an individual to have multiple locations and each location for the individual to have specific attributes like date_time, altitude, relative_position and notes.
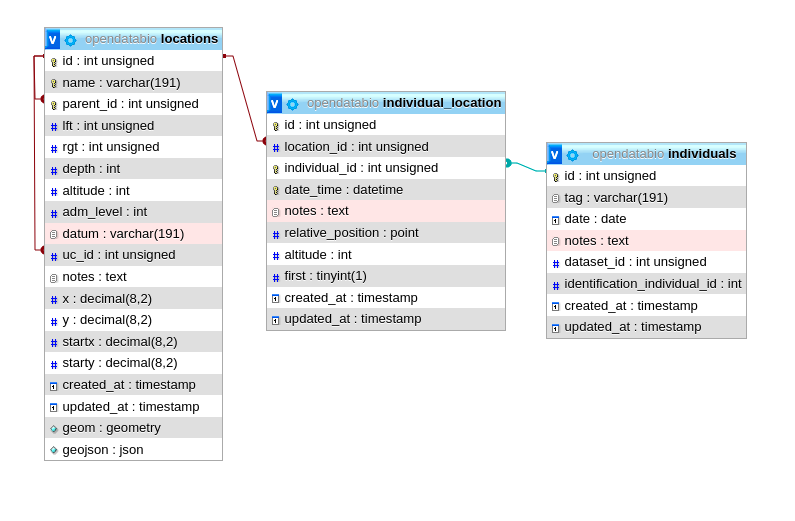 The same tables related with the Location model with the direct and non-polymoprhic relationships indicated.
The same tables related with the Location model with the direct and non-polymoprhic relationships indicated.
Location Table Columns
- Columns
parent_id together with rgt, lft and deph are used to define the Nested Set Model to query ancestors and descendants in a fast way. Only parent_id is specified by the user, the other columns are calculated by the Baum library trait from the id+parent_id values that define the hierarchy. The same hierarchical model is used for the Taxon Model, but for Locations there is a spatial constraint, i.e. a children must fall within a parent geometry. - The
adm_level column indicate the administrative level, or type, of a location. By default, the following adm_level are configured in OpenDataBio:2 for country, 3 for first division within country (province, state), 4 for second division (e.g. municipality),… up to adm_level=10 as administrative areas (country code is 2 to allow standardization with OpenStreeMaps, which is recommended to follow if your installation will include data from different countries). The administrative levels may be configured in an OpenDataBio before importing any data to the database, see the installation guide for details on that.99 is the code for Conservation Units - a conservation unit is a location that may be linked to multiple other locations (any location may belong to a single UC). Thus, one Location may have as parent a city and as uc_id the conservation unit where it belongs.98 is the code for Indigenous Territories - same properties as Conservation Units, but treated separately only because some CUs and TIs may largely overlap as is the case the Amazon region97 ise the code for Environmental layers - same properties as Conservation Units and Indigenous Territories, i.e., may be linked as additional location to any Point, Plot or Transect, and thehence, their related individuals. Store polygons and multipolygon geometries representing environmental classes, such as vegetation units, biomes, soil classes, etc…100 is the code for plots and subplots - plot locations may be registered with Point or with a Polygon geometry, and must also have an associated Cartesian dimensions in meters. If it is a point location, the geometry is defined by ODB from the dimensions with NorthEast orientation from the point informed. Cartesian dimensions of a plot location can also be combined with cartesian positions of subplots (i.e. a plot location whose parent is also a plot location) and/or of individuals within such plots, allowing individuals and subplots to be mapped within a plot subplot location without geometry specifications. In other words, if the spatial geometry of the plot is unknown, it may have as geometry a single GPS point rather than a polygon, plus its x and y dimensions. A subplot is location plot inside a location plot and must consist of a point marking the start of the subplot plus its X and Y cartesian dimensions. If the geometry of the start of the subplot is unknown, it may be stored as a relative position to parent plot using the startx and starty.101 for transects - like plots, transects may be registered having a LineString geometry or simply a single Latitude and Longitude coordinates and a dimension. The x cartesian dimension for transects represent the length in meters and is used to create a linestring (North oriented) when only a point is informed. The y dimension is used to validate individuals as belonging to transect location, and represents the maximum distance from the line that and individual must fall to be detected in that location.999 for ‘POINT’ locations like GPS waypoints - this is for registration of any point in space
- Column
datum may record the geometry datum property, if known. If left blank, the location is considered to be stored using WGS84 datum. However, there is no built-in conversor from other types of data, so the maps displayed may be incorrect if different datum’s are used. Strongly recommended to project data as WSG84 for standardization. - Column
geom stores the location geometry in the database, allowing spatial queries in SQL language, such as parent autodetection. The geometry of a location may be POINT, POLYGON, MULTIPOLYGON or LINESTRING and must be formatted using Well-Known-Text geometry representation of the location. When a POLYGON is informed, the first point within the geometry string is privileged, i.e. it may be used as a reference for relative markings. For example, such point will be the reference for the startx and starty columns of a subplot location. So for plot and transect geometries, it matters which point is listed first in the WKT geometry
Data access Full users may register new locations, edit locations details and remove locations records that have no associated data. Locations have open access!
Individual Model
The Individual object represents a record for an individual organism. It may be a single time-space occurrence of an animal, plant or fungi, or an individual monitored through time, such as a plant in a permanent forest plot, or an animal in capture-recapture or radio-tracking experiment.
An Individual may have one or more Vouchersrepresenting physical samples of the individual stored in one or more Biological Collection and it may have one or more Locations, representing the place or places where the individual has been recorded.
Individual objects may also have a self taxonomic Identification or its taxonomic identity may depend on that of another individual (non-self identification). The Individual identification is inherited by all the Vouchers registered for the Individual. Hence Vouchers do not have their separate identification.
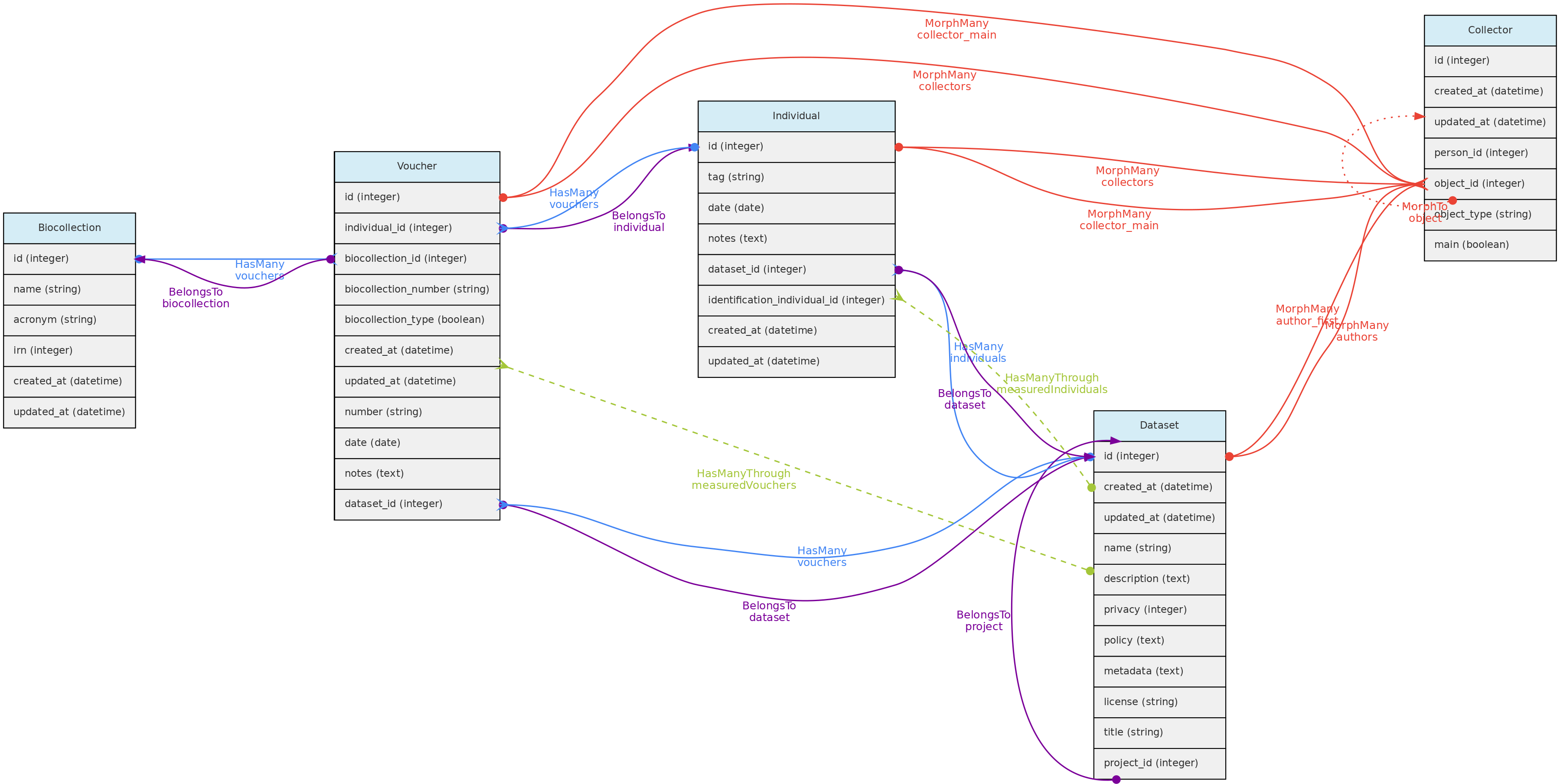
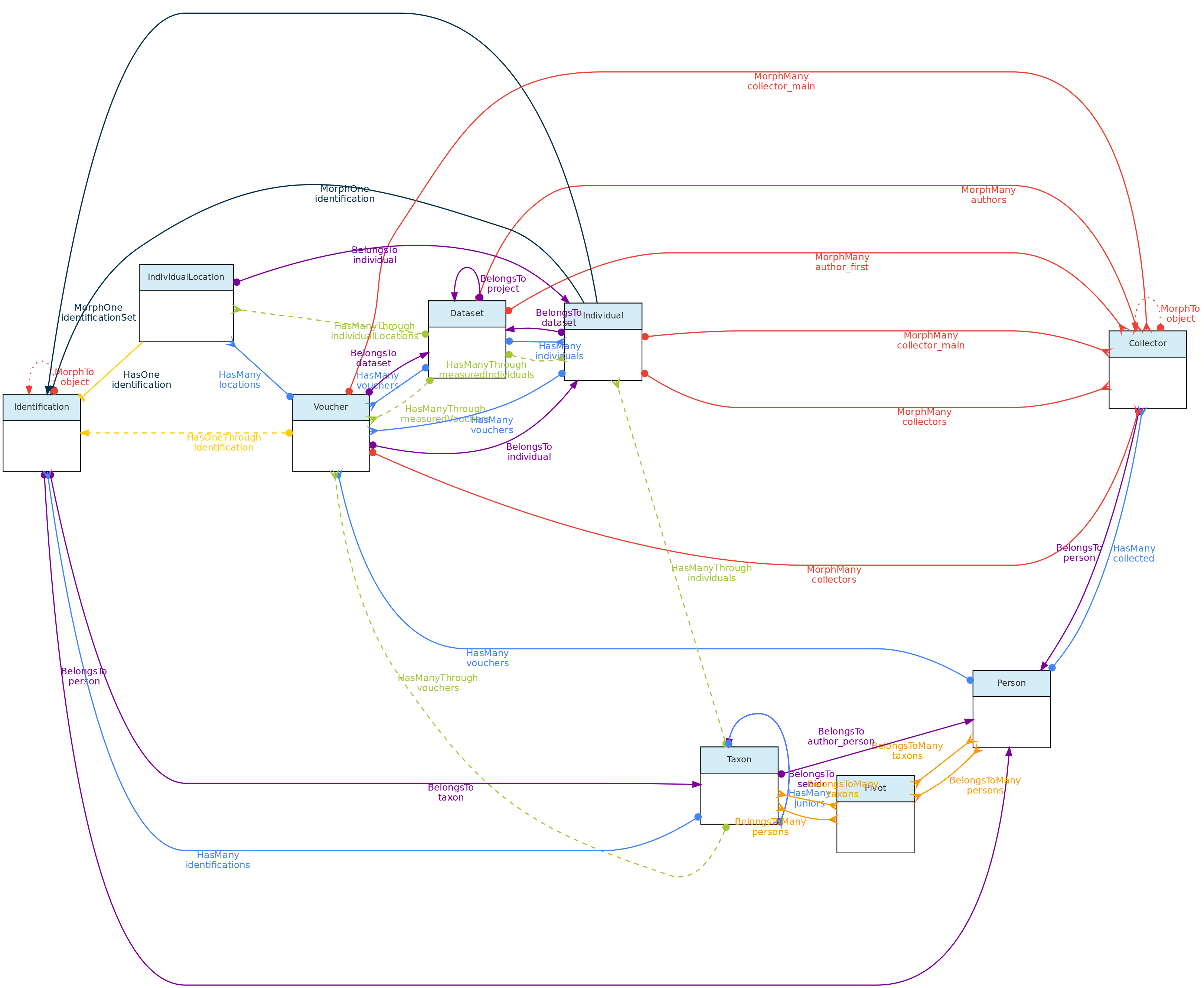 This figure shows the Individual Model and the models it relates to, except the Measurement and Location models, as their relationships with Individuals is shown elsewhere in this page. Lines linking models indicate the
This figure shows the Individual Model and the models it relates to, except the Measurement and Location models, as their relationships with Individuals is shown elsewhere in this page. Lines linking models indicate the methods or functions implemented in the classes to access the relationships. Dashed lines indicate indirect relationships and the colors the different types of Laravel Eloquent methods.
 The Individual model direct and non-polymoprhic relationships.
The Individual model direct and non-polymoprhic relationships.
Individual Table Columns
- A Individual record must specify at least one Location where it was registered, the
date of registration, the local identifier tag, and the collectors of the record, and the dataset_id the individual belongs to. - The Location may be any location registered, regardless of level, allowing to store historical records whose georeferencing is just an administrative location. Individual locations are stored in the
individual_location pivot table, having columns date_time, altitude, notes and relative_position for the individual location records. - The column
relative_position stores the Cartesian coordinates of the Individual in relation to its Location. This is only for individuals located in locations of type plot, transect or point. For example, a Plot location with dimensions 100x100 meters (1ha) may have an Individual with relative position=POINT(50 50), which will place the individual in the center of the location (this is shown graphically in the web-interface), as is defined by the x and y coordinates of the individual. If the location is a subplot, then the position within the parent plot may also be calculated (this was designed with ForestGeo plots in mind and is a column in the Individual GET API. If the location is a POINT, the relative_position may be informed as angle (= azimuth) and distance, attributes frequently measured in sampling methods. If the location is a TRANSECT, the relative_position places the individual in relation to the linestring, the x being the distance along the transect from the first point, and the y the perpendicular distance where the individual is located, also accounting for some sampling methods; - The
date field in the Individual, Voucher, Measurement and Identification models may be an Incomplete Date, i.e., only the year or year+month may be recorded. - The Collector table represents collectors for an Individual or Voucher, and is linked with the Person Model. The collector table has a polymorphic relationship with the Voucher and Individual objects, defined by columns
object_id and object_type, allowing multiple collectors for each individual or voucher record. The main_collector indicated is just the first collector listed for these entities. - The
tag field is a user code or identifier for the Individual. It may be the number written on the aluminum tag of a tree in a forest plot, the number of a bird-band, or the collector number of a specimen. The combination of main_collector+tag+first_location is constrained to be unique in OpenDataBio. - The taxonomic identification of an Individual may be defined in two ways:
- for self identifications an Identification record is created in the identifications table, and the column
identification_individual_id is filled with the Individual own id - for non-self identifications, the id of the Individual having the actual Identification is stored in column
identification_individual_id. - Hence, the Individual class contain two methods to relate to the Identification model: one that sets self identifications and another that retrieves the actual taxonomic identifications by using column
identification_individual_id.
- Individuals may have one or more Vouchersdeposited in a Biocollection.
Data access Individuals belong to Datasets, so Dataset access policy apply to the individuals in it. Only project collaborators and administrators may insert or edit individuals in a dataset, even if dataset is of public access.
Taxon Model
The general idea behind the Taxon model is to present tools for easily incorporating valid taxonomic names from Online Taxonomic Repositories (currently Tropicos.org and GBIF are implemented), but allowing for the inclusion of names that are not considered valid, either because they are still unpublished (e.g. a morphotype), or the user disagrees with published synonymia, or the user wants to have all synonyms registered as invalid taxons in the system. Moreover, it allows one to define a custom clade level for taxons, allowing one to store, in addition to taxonomic rank categories, any node of the tree of life. Any registered Taxon can be used in Individual identifications and Measurements may be linked to taxonomic names.
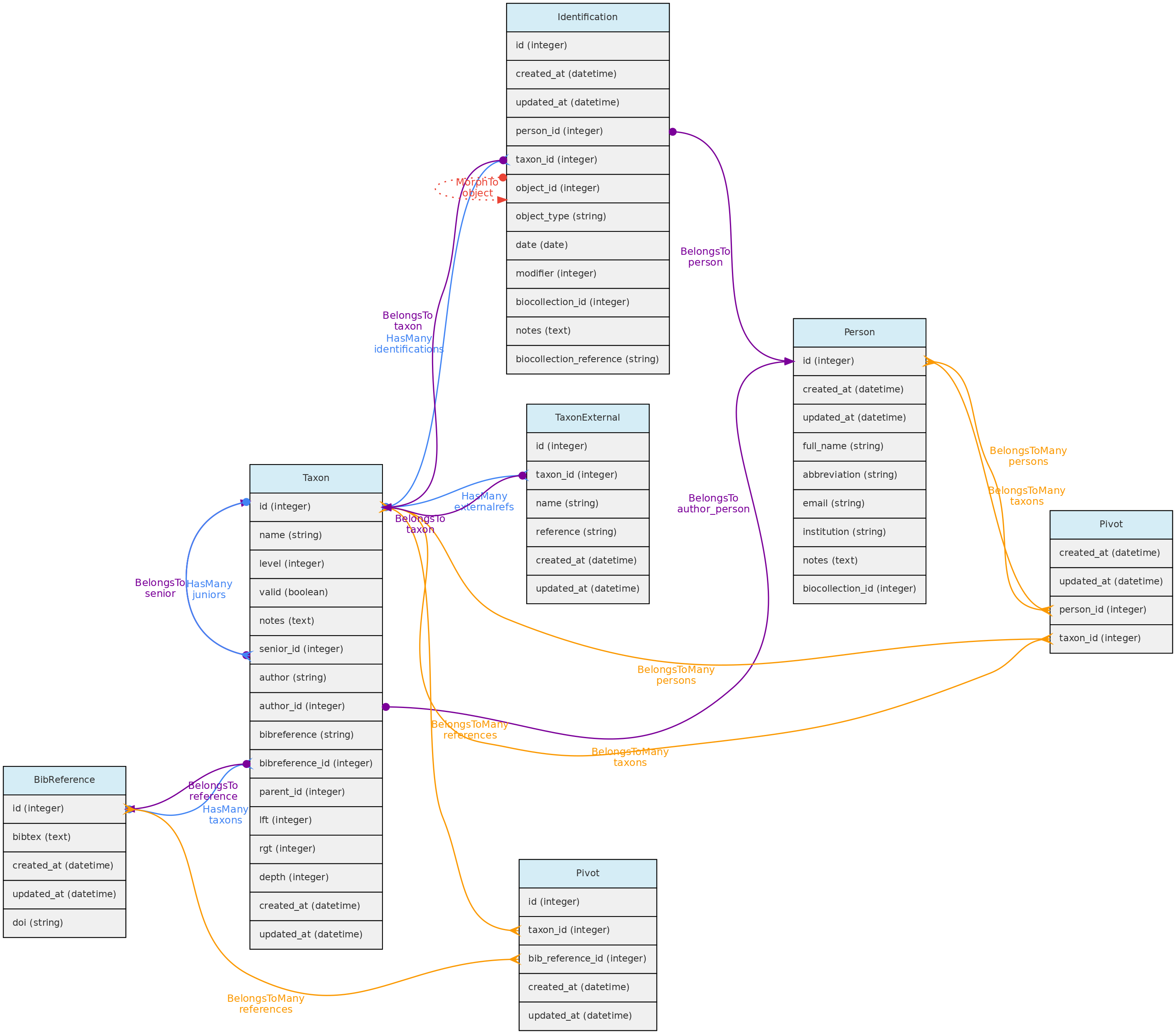 Taxon model and its relationships. Lines linking tables indicate the
Taxon model and its relationships. Lines linking tables indicate the methods implemented in the shown classes, with colors indicating different Eloquent relationships
Taxon table explained
- Like, Locations, the Taxon model has a parent-child relationship, implemented using the Nested Set Model for hierarchical data of the Laravel library Baum that allows to query ancestors and descendants. Hence, columns
rgt, lft and deph of the taxon table are automatically filled by this library upon data insertion or update. - For both, Taxon
author and Taxon bibreference there are two options:- For published names, the string authorship retrieved by the external taxon APIs will be placed in the
author=string column. For unpublished names, author is a Person and will be stored in the author_id column. - Only published names may have relation to BibReferences. The
bibreference string field of the Taxon table stores the strings retrieved through the external APIs, while the bibreference_id links to a BibReference object. These are used to store the Publication where the Taxon Name is described and may be entered in both formats. - In addition, a Taxon record may also have many other BibReferences through a pivot table (
taxons_bibreference), permitting to link any number of bibliographic references to a Taxon name.
- Column
level represents the taxonomic rank (such as order, genera, etc). It is numerically coded and standardized following the IAPT general rules, but should accommodate also animal related taxon level categories. See the available codes in the Taxon API for the list of codes. - Column
parent_id indicates the parent of the taxon, which may be several levels above it. The parent level should be strictly higher than the taxon level, but you do not need to follow the full hierarchy. It is possible to register a taxon without parents, for example, an unpublished morphotype for which both genera and family are unknown may have an order as parent. - Names for the taxonomic ranks are translated according to the system defined
locale that also translates the web interface (currently only Portuguese and English implemented). - The
name field of the taxon table contain only the specific part of name (in case of species, the specific epithet), but the insertion and display of taxons through the API or webinterface should be done with the fullname combination. - It is possible to include synonyms in the Taxon table. To do so, one must fill in the
senior relationship, which is the id of the accepted (valid) name for an invalid Taxon. If senior_id is filled, then the taxon is a junior synonym and must be flagged as invalid. - When inserting a new published taxon, only the
name is required. The name will be validated and the author, reference and synonyms will be retrieved using the following API services:- GBIF BackBone Taxonomy - this will be the first check, from which links to Tropicos and IPNI may also be retrieved if registering a plant name.
- Tropicos - if not found on GBIF, ODB will search the name on the Missouri Botanical Garden nomenclature database.
- IPNI - the International Individual Names Index is another database used to validate individual names (Temporarily disabled)
- MycoBank - used to validate a name if not found by the Tropicos nor IPNI apis, and used to validate names for Fungi. Temporarily disabled
- ZOOBANK - when GBIF, Tropicos, IPNI and MycoBank fails to find a name, then the name is tested against the ZOOBANK api, which validates animal names. Does not provide taxon publication, however.
- If a Taxon name is found in the Nomenclatural databases, the respective ID of the repository is stored in the
taxon_external tables, creating a link between the OpenDataBio taxon record and the external nomenclatural database. - A Person may be defined as one or more taxon specialist through a pivot table. So, a Taxon object may have many taxonomic specialist registered in OpenDataBio.
Data access: Full users are able to register a new taxon and edit existing records if they have not been used for Identification of Measurements. Currently it is impossible to remove a taxon from the database. Taxon list have public access.
Voucher Model
The Voucher model is used to store records of specimens or samples from Individuals deposited in Biological Collections. Therefore, the only mandatory information required to register a Voucher are individual, biocollection and whether the specimen is a nomenclature type (which defaults to non-type if not informed).
 Voucher model and its relationships. Lines linking tables indicate the
Voucher model and its relationships. Lines linking tables indicate the methods implemented in the shown models, with colors indicating different Eloquent relationships. Not that Identification nor Location are show because Vouchers do not have their own records for these two models, they are just inherited from the Individual the Voucher belongs to
Vouchers table explained
- A Voucher belongs to an Individual and a Biocollection, so the
individual_id and the biocollection_id are mandatory in this table; biocollection_number is the alpha-numeric code of the Voucher in the BioCollection, it may be ’null’ for users that just want to indicate that a registered Individual have Vouchers in a particular Bicollection, or to registered Vouchers for biocollections that do not have an identifier code;biocollection_type - is a numeric code that specify whether the Voucher in the BioCollection is a nomenclatural type. Defaults to 0 (Not a Type); 1 for just ‘Type’, a generic form, and other numbers for other nomenclature type names (see the API Vouchers Endpoint for a full list of options).collectors, one or multiple, are optional for Vouchers, required only if they are different from the Individual collectors. Otherwise the Individual collectors are inherited by the Voucher. Like for Individuals, these are implemented through a polymorphic relationship with the collectors table and the first collector is the main_collector for the voucher, i.e. the one that relates to number.number, this is the collector number, but like collectors, should only be filled if different from the Individual’s tag value. Hence, collectors, number and date are useful for registering Vouchers for Individuals that have Vouchers collected at different times by different people.date field in the Individual and Voucher models may be an incomplete date. Only required if different from that of the Individual the Voucher belongs to.dataset_id the Voucher belongs to a Dataset, which controls the access policy;notes any text annotation for the Voucher.- The Voucher model interacts with the BibReference model, permitting to link multiple citations to Vouchers. This is done with a pivot
voucher_bibreference table.
Data access Vouchers belong to Datasets, so Dataset access policy apply to the Vouchers in it. Vouchers may have a different Project than their Individuals. If the Voucher dataset policy is open access and that of the Individual project is not, then access to voucher data will be incomplete, so Voucher’s dataset should have the same or less restricted access policy than the Individual dataset. Only Dataset collaborators and administrators may insert or edit vouchers in a dataset, even if the dataset is of public access.
2 - Trait Objects
Objects for user defined variables and their measurements
Measurement Model
The Measurements table stores the values for traits measured for core objects. Its relationship with the core objects is defined by a polymorphic relationship using columns measured_id and measured_type. These MorphTo relations are illustrated and explained in the core objects page.
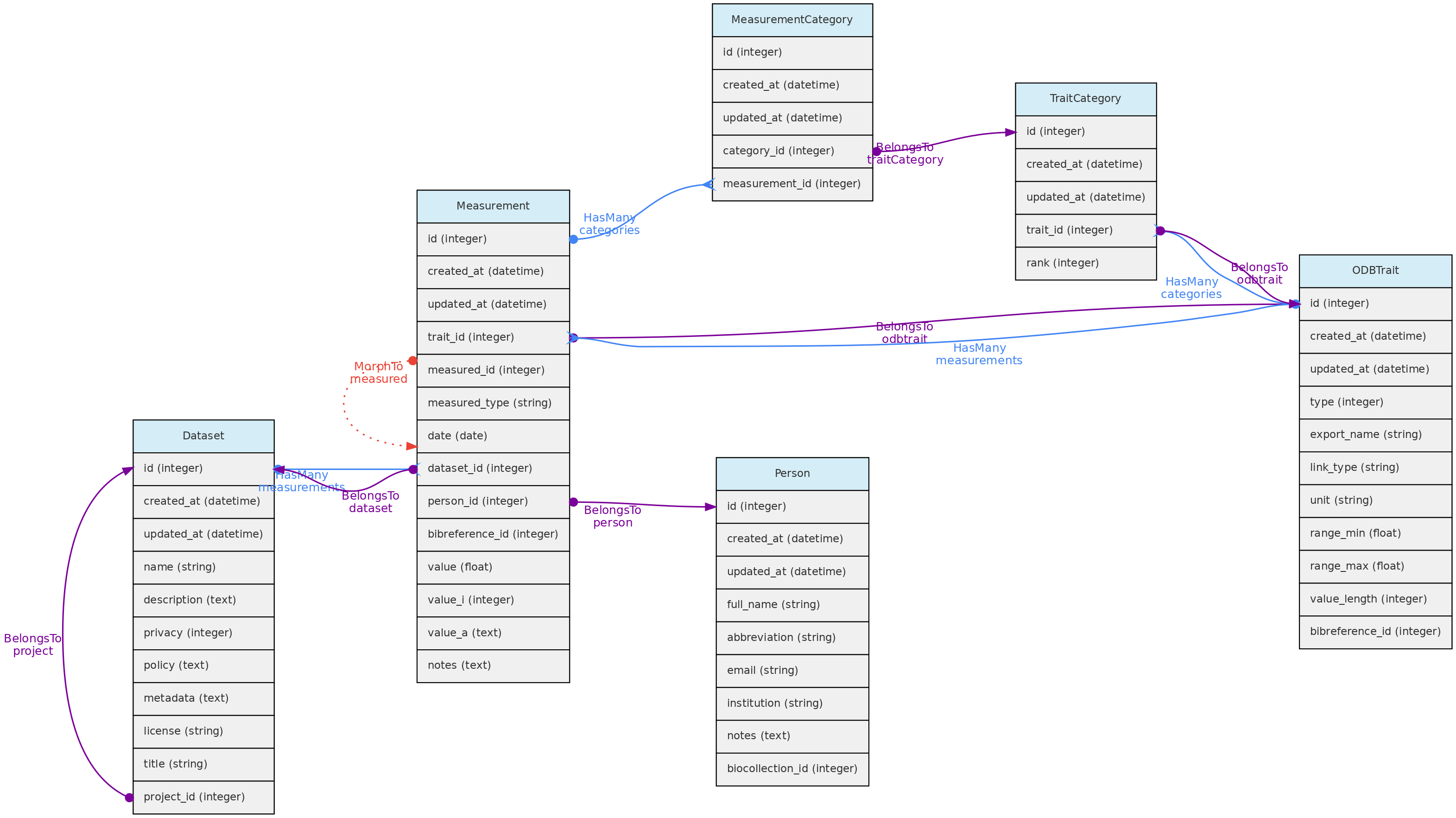
- Measurements must belong to a Dataset - column
dataset_id, which controls measurement access policy - A Person must be indicated as a measurer (
person_id); - The
bibreference_id column may be used to link measurements extracted from publications to its Bibreference source; - The value for the measured trait (
trait_id) will be stored in different columns, depending on trait type:value - this float column will store values for Quantitative Real traits;value_i - this integer column will store values for Quantitative Integer traits; and is an optional field for Link type traits, allowing for example to store counts for a species (a Taxon Link trait) in a location.value_a - this text column will store values for Text, Color and Spectral trait types.
- Values for Categorical and Ordinal traits are stored in the
measurement_category table, which links measurements to trait categories. date - measurement date is mandatory in all cases
Data access Measurements belong to Datasets, so Dataset access policy apply to the measurements in it. Only dataset collaborators and administrators may insert or edit measurements in a dataset, even if the dataset is of public access.
Trait Model
The ODBTrait table represents user defined variables to collect Measurements for one of the core object, either Individual, Voucher, Location or Taxon.
These custom traits give enormous flexibility to users to register their variables of interest. Clearly, such flexibility has a cost in data standardization, as the same variable may be registered as different Traits in any OpenDataBio installation. To minimize redundancy in trait ontology, users creating traits are warned about this issue and a list of similar traits is presented in case found by trait name comparison.
Traits have editing restrictions to avoid data loss or unintended data meaning change. So, although the Trait list is available to all users, trait definitions may not be changed if somebody else also used the trait for storing measurements.
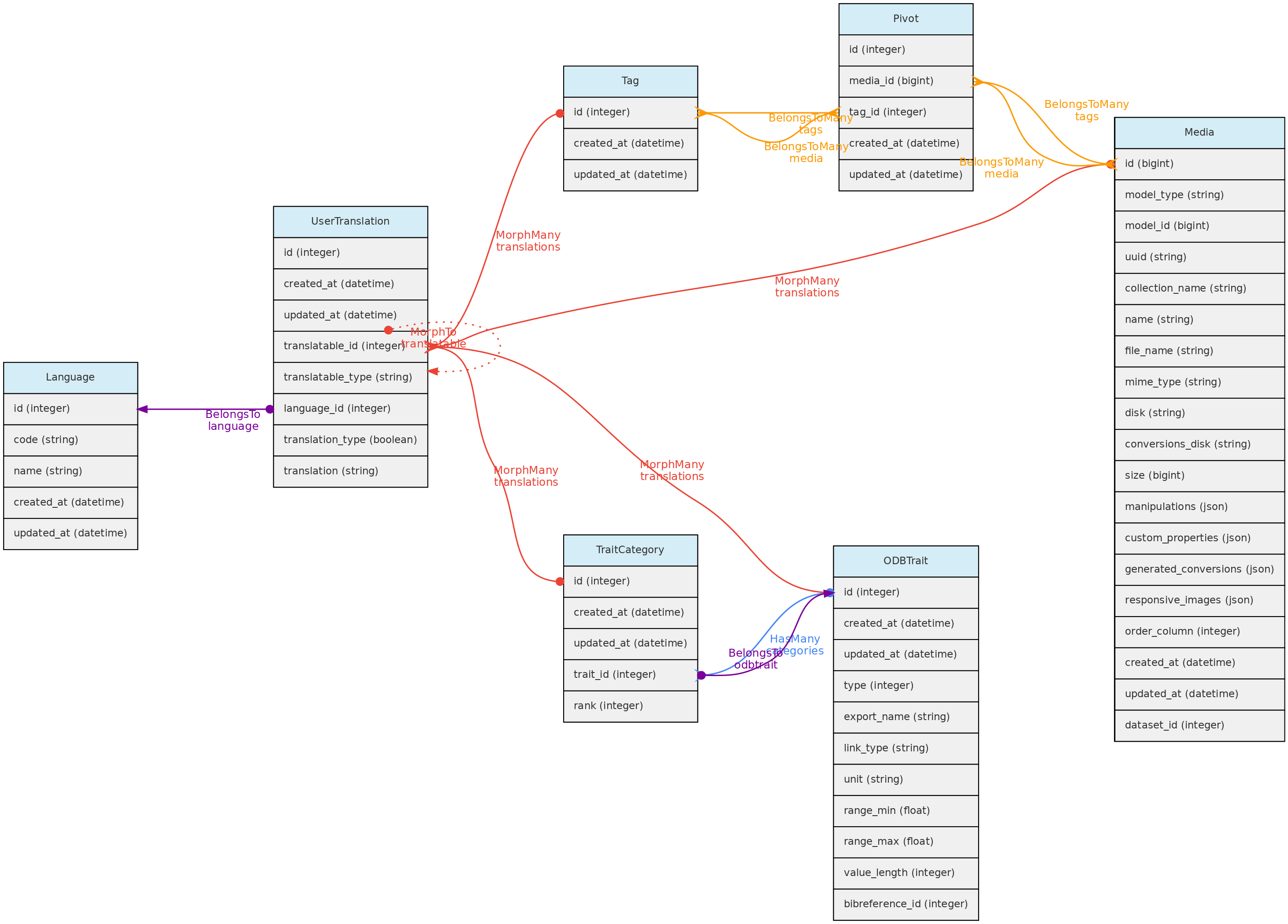
Traits are translatable entities, so their name and description values can be stored in multiple languages (see User Translations. This is placed in the user_translations table through a polymorphic relationship.
The Trait definition should be as specific as needed. The measurement of tree heights using direct measurement or a clinometer, for example, may not be easily converted from each other, and should be stored in different Traits. Thus, it is strongly recommended that the Trait definition field include information such as measurement instrument and other metadata that allows other users to understand whether they can use your trait or create a new one.
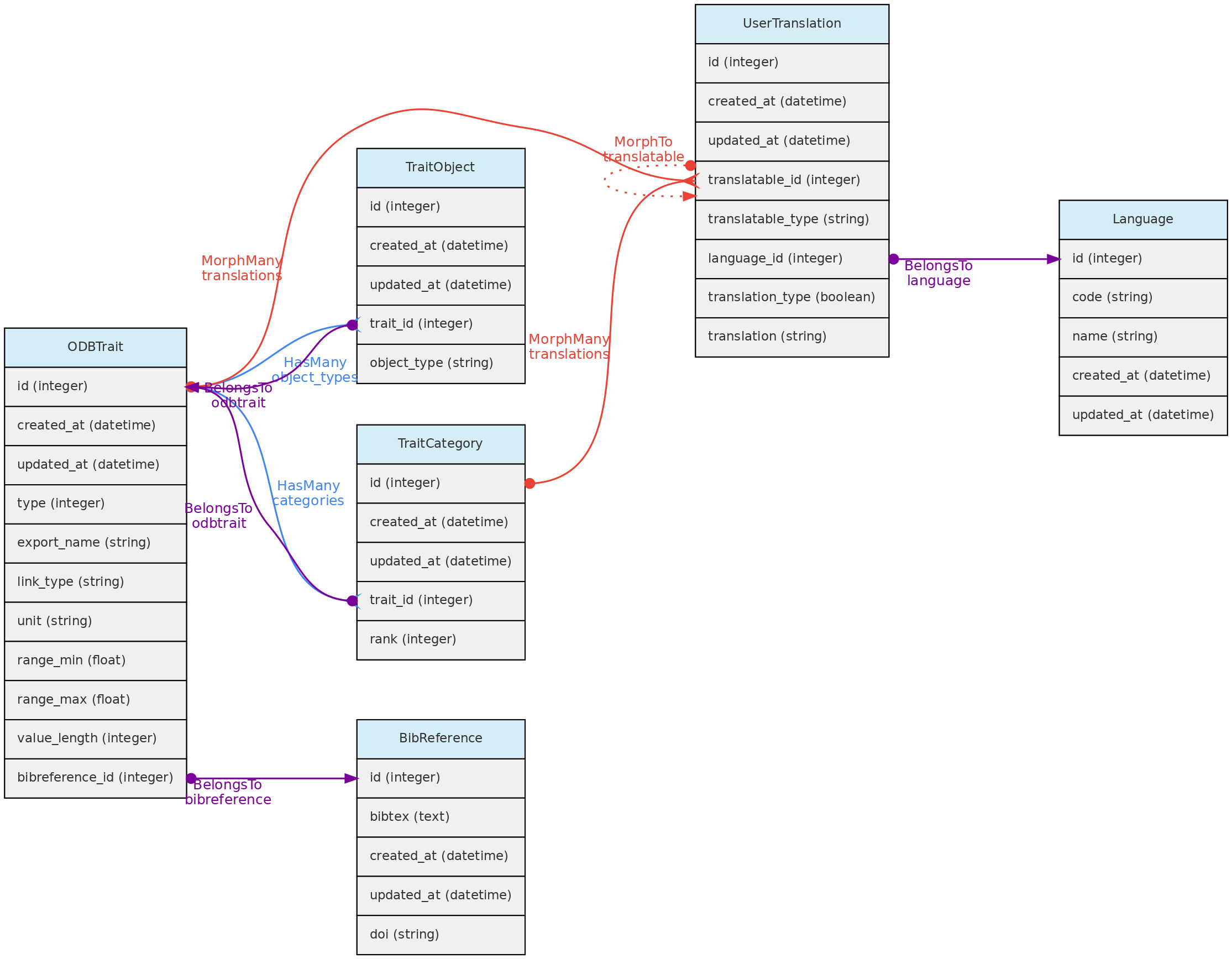
- The Trait definition must include an
export_name for the trait, which will be used during data exports and are more easily used in trait selection inputs in the web-interface. Export names must be unique and have no translation. Short and camelCase or PascalCase export names are recommended. - The following trait types are available:
- Quantitative real - for real numbers;
- Quantitative integer - for counts;
- Categorical - for one selectable categories;
- Categorical multiple - for many selectable categories;
- Categorical ordinal - for one selectable ordered categories (semi-quantitative data);
- Text - for any text value;
- Color - for any color value, specified by the hexadecimal color code, allowing renderizations of the actual color.
- Link - this is a special trait type in OpenDataBio to link to database object. Currently, only link to Taxons and Voucher are allowed as a link type traits. Use ex: if you want to store species counts conducted in a location, you may create a Taxon link type Trait or a Voucher link type Trait if the taxon has vouchers. A measurement for such trait will have an optional
value field to store the counts. This trait type may also be used to specify the host of a parasite, or the number of predator insects. - Spectral - this is designed to accommodate Spectral data, composed of multiple absorbance or reflectance values for different wavenumbers.
- GenBank - this stores GenBank accessions numbers allowing to retrieve molecular data linked to individuals or vouchers stored in the database through the GenBank API Service.
- The Traits table contains fields that allow measurement value validation, depending on trait type:
range_max and range_min - if defined for Quantitative traits, measurements will have to fit the specified range;value_length - mandatory for Spectral Traits only, validate the length (number of values) of a spectral measurement;link_type - if trait is Link type, the measurement value_i must be an id of the link type object;- Color traits are validated in the measurement creation process and must conform to a color hexadecimal code. A color picker is presented in the web interface for measurement insertion and edition;
- Categorical and ordinal traits will be validated for the registered categories when importing measurements through the API;
- Column
unit defines the measurement unit for the Trait. There is no way to prevent measurements values imported with a distinct unit. Quantitative traits required unit definition. - Column
bibreference_id is the key of a single BibReference that may be linked to trait definition. - The
trait_objects table stores the type of core object (Taxon, Location, Voucher) that the trait can have a measurement for;
Data access A Trait name, definition, unit and categories may not be updated or removed if there is any measurement of this trait registered in the database. The only exceptions are: (a) it is allowed to add new categories to categorical (not ordinal) traits; (b) the user updating the trait is the only Person that has measurements for the trait; (c) the user updating the trait is an Admin of all the datasets having measurements using trait.
A Form is an organized group of Traits, defined by a User in order to create a custom form that can be filled in for entering measurements through the web interface. A Form consists of a group of ordered Traits, which can be marked as “mandatory”. Related entities are the Report and the Filter.
This is still experimental and needs deeper testing
3 - Data Access Objects
Objects controlling data access and distribution!
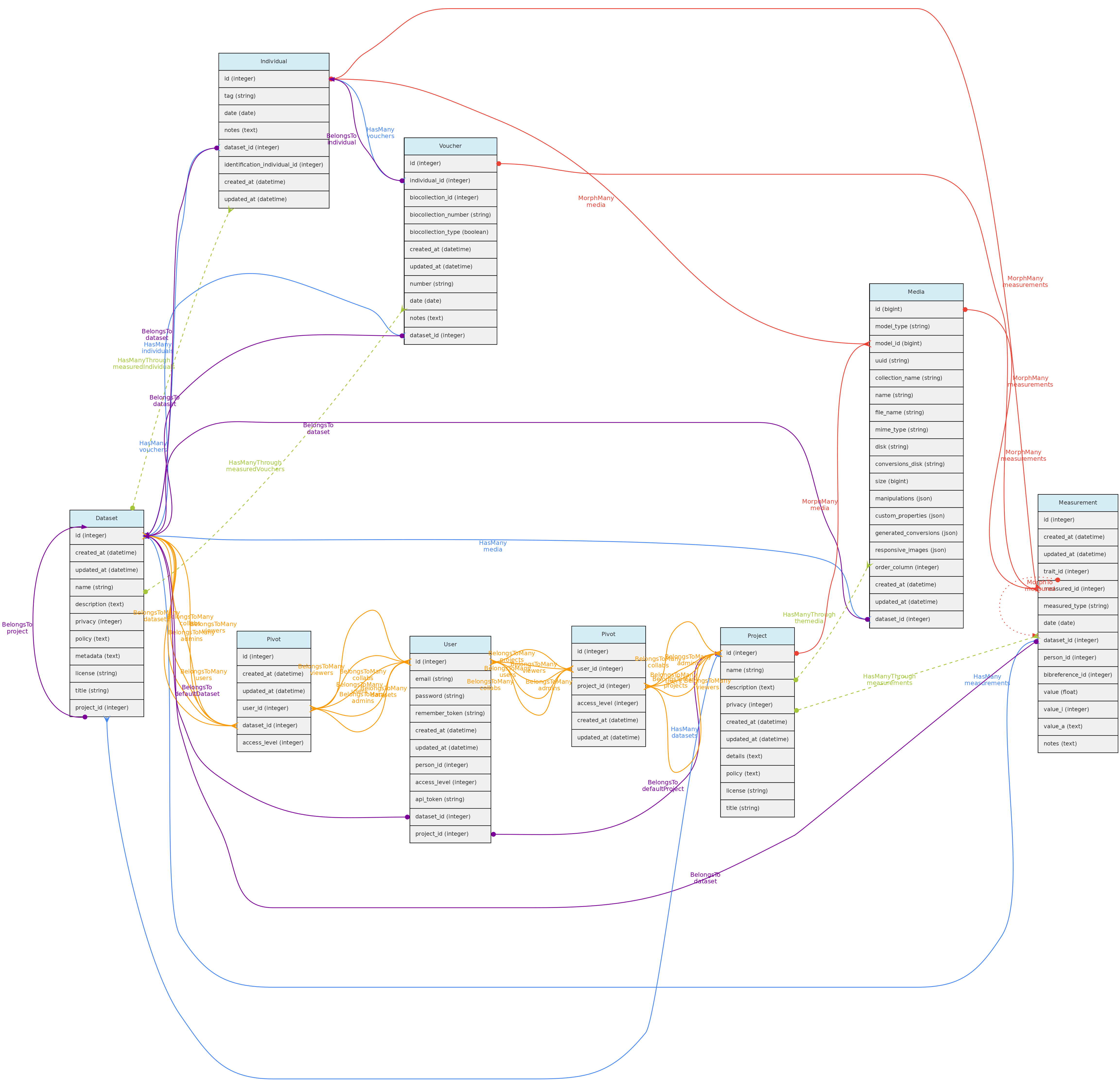
Datasets control data access and represents a dynamic data publication, with a version defined by last edition date. Datasets may contain Measurements, Individuals, Vouchers and/or Media Files.
Projects are just groups of Datasets and Users, representing coohorts of users with common accessibility to datasets whose privacy are set to be controlled by a Project.
BioCollections - This model serves to create a reusable list of acronyms of Biological Collections to record Vouchers. However, you can optionally manage a collection of Vouchers and their Individuals, in parallel to the access control provided by Datasets. Control is only for editing and entering Voucher records. In this case, the BioCollection is managed by the system.
Projects and BioCollections must have at least one User defined as administrator, who has total control over the dataset or project, including granting the following roles to other users: administrator, collaborator or viewer:
- Collaborators are able to insert and edit objects, but are not able to delete records nor change the dataset or project configuration.
- Viewers have read-only access to the data that are not of open access.
- Only Full Users and SuperAdmins may be assigned as administrators or collaborators. Thus, if a user who was administrator or collaborator of a dataset is demoted to “Registered User”, she or he will become a viewer.
- Only Super-admins can enable a BioCollection to be administered by the system.
Biocollections
The Biocollection model has two functions: (1) to provide a list of acronyms for registering Vouchers of any Biological Collection; (2) to manage the data of Biological Collections, facilitating the registration of new data (any user enters their data using the validations carried out by the software and requests the collection’s curators through the interface to register the data, which is done by authorized users for the BioCollection. Upon data registration, the BioCollection controls the editing of the data of the Vouchers and the related Individuals. Option (2) needs to be implemented by a Super-Administrator user, who can enable a BioCollection to be administered by the system, implementing the ODBRequest request model so that users can request data, samples, or records and changes to the data.
The Biocollection object may be a formal Biocollection, such as those registered in the Index Herbariorum (http://sweetgum.nybg.org/science/ih/), or any other Biological Collection, formal or informal.
The Biocollection object also interacts with the Person model. When a Person is linked to an Biocollection it will be listed as a taxonomic specialist.
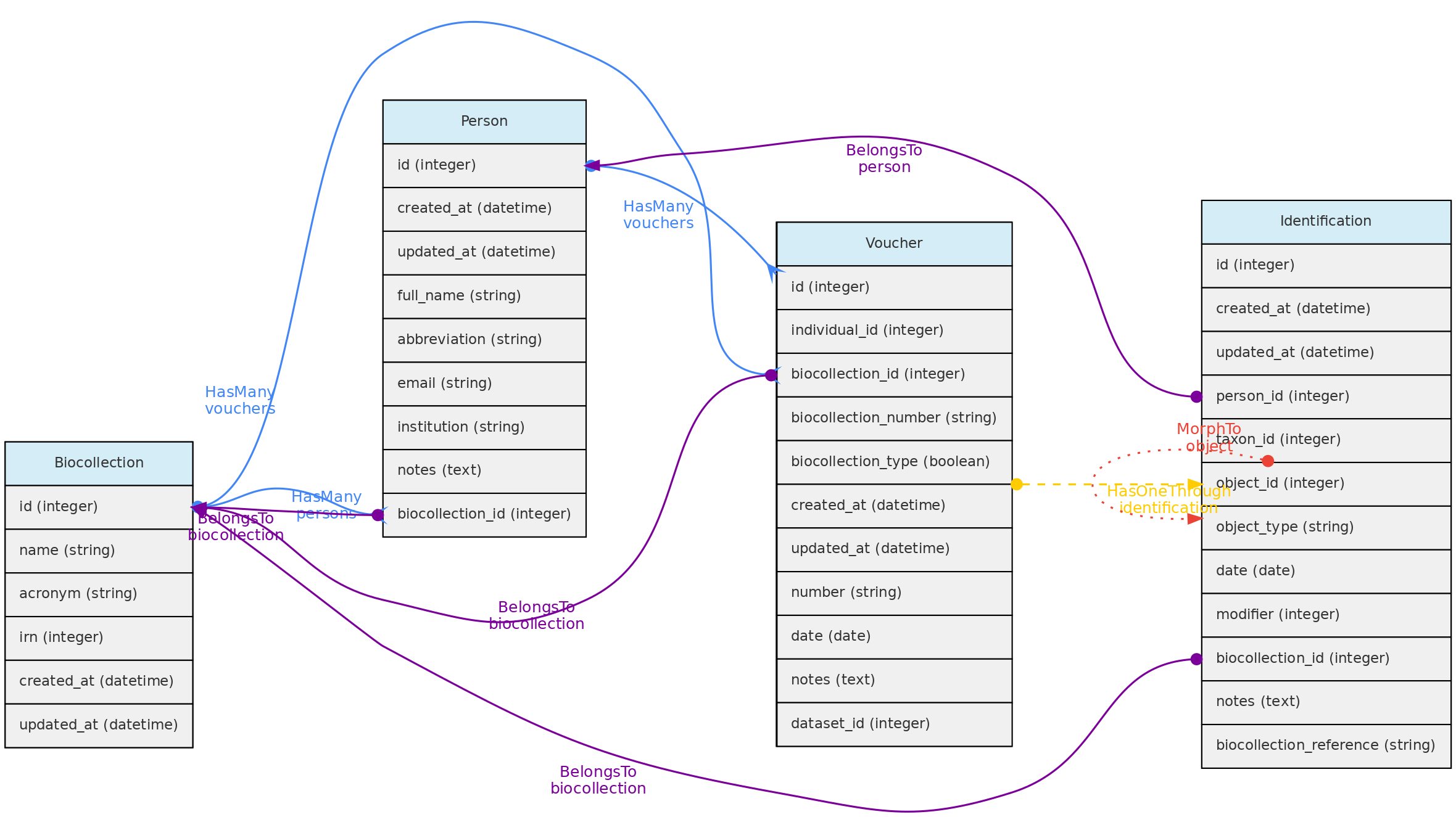
Data access - Full users can register BioCollections, but only super administrator can make a BioCollection manageable by the system. Removing BioCollections can be done if there are no Vouchers attached and if it is not administered by the system. If manageable, the model interacts with Users, which can be administrators (curators, anything can) or collaborators (can enter and edit data, but cannot delete records). Data from other Datasets can be part of the BioCollection, allowing users to have their complete data, but editing control of records is by the BioCollection authorized users.
Datasets
DataSets are groups of Measurements, Individuals, Vouchers and/or Media Files, and may have one or more Users administrators, collaborators or viewers. Administrators may set the privacy level to public access, restricted to registered users or restricted to authorized users or restricted to project users. This control access to the data within a dataset as exemplified in diagram below:
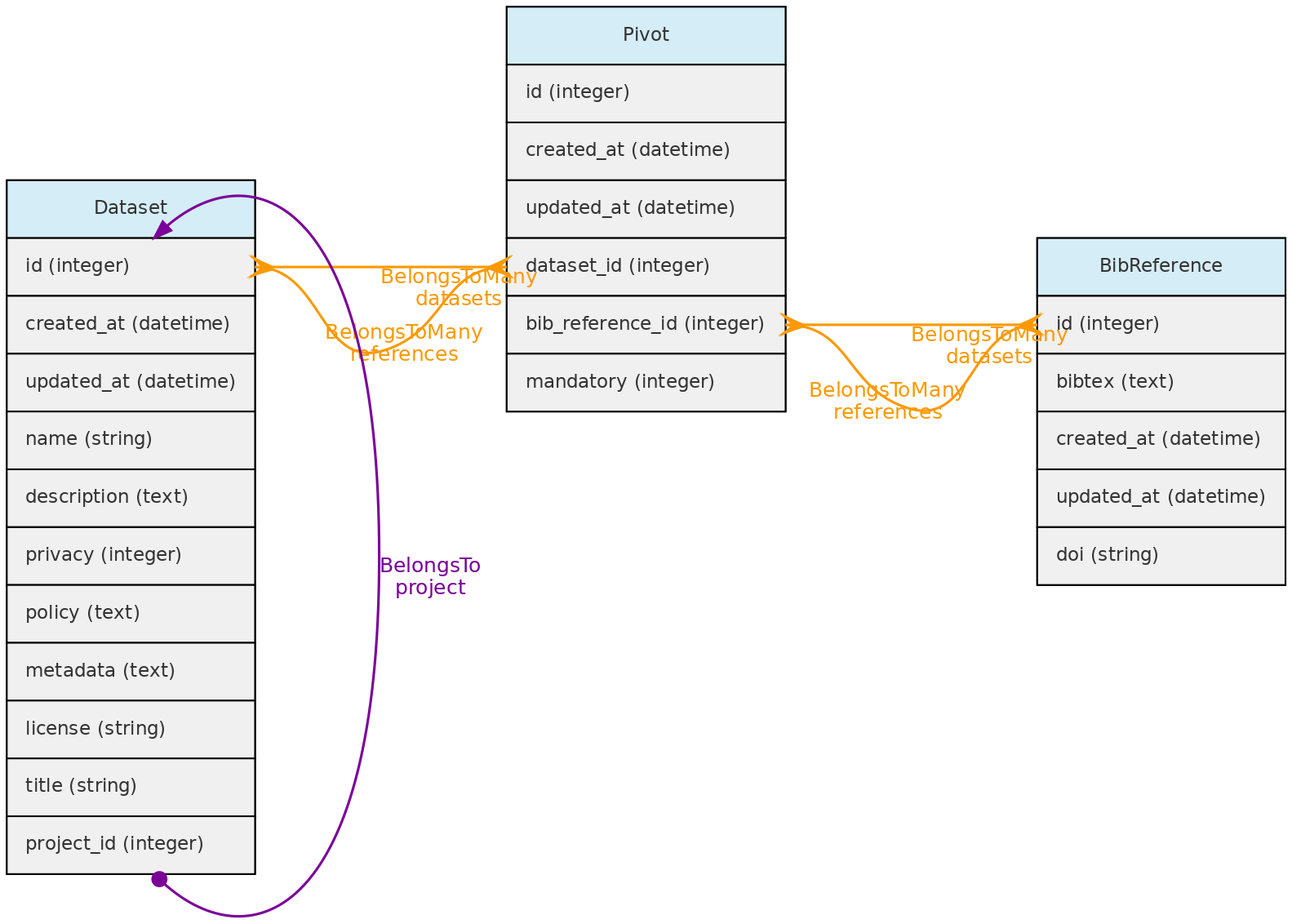
Datasets may also have many Bibliographic References, which together with fields policy, metadata permits to annotate the dataset with relevant information for data sharing:
* Link any publication that have used the dataset and optionally indicate that they are of mandatory citation when using the data;
* Define a specific data policy when using the data in addition to the a CreativeCommons.org public license;
* Detail any relevant metadata in addition to those that are automatically retrieved from the database like the definitions of the Traits measured.
Projects
Projects are just groups of Datasets and interacts with Users, having administrators, collaborators or viewers. These users may control all datasets within the Project having a restricted to project users access policy.
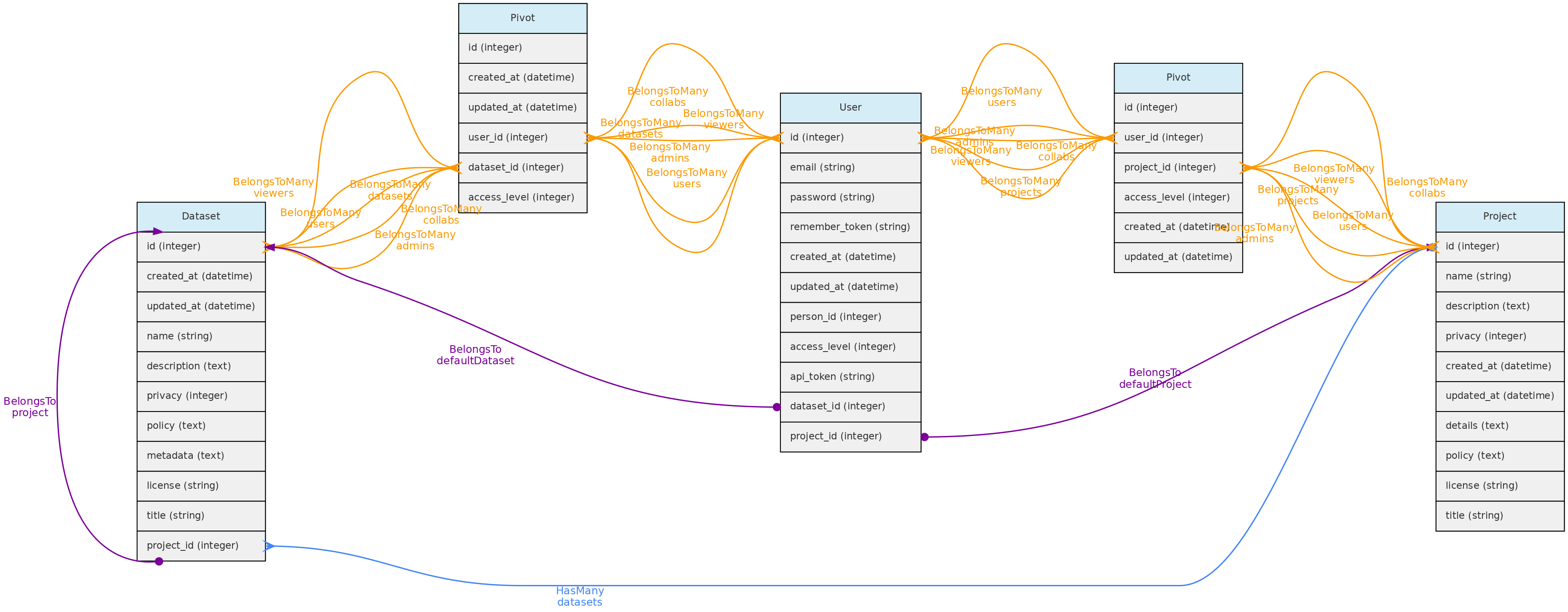
Users
The Users table stores information about the database users and administrators. Each User may be associated with a default Person. When this user enters new data, this person is used as the default person in forms. The person can only be associated to a single user.
There are three possible access levels for a user:
* Registered User (the lowest level) - have very few permissions
* Full User - may be assigned as administrators or collaborators to Projects and Datasets;
* SuperAdmin (the highest level). - superadmins have have access to all objects, regardless of project or dataset configuration and is the system administrator.
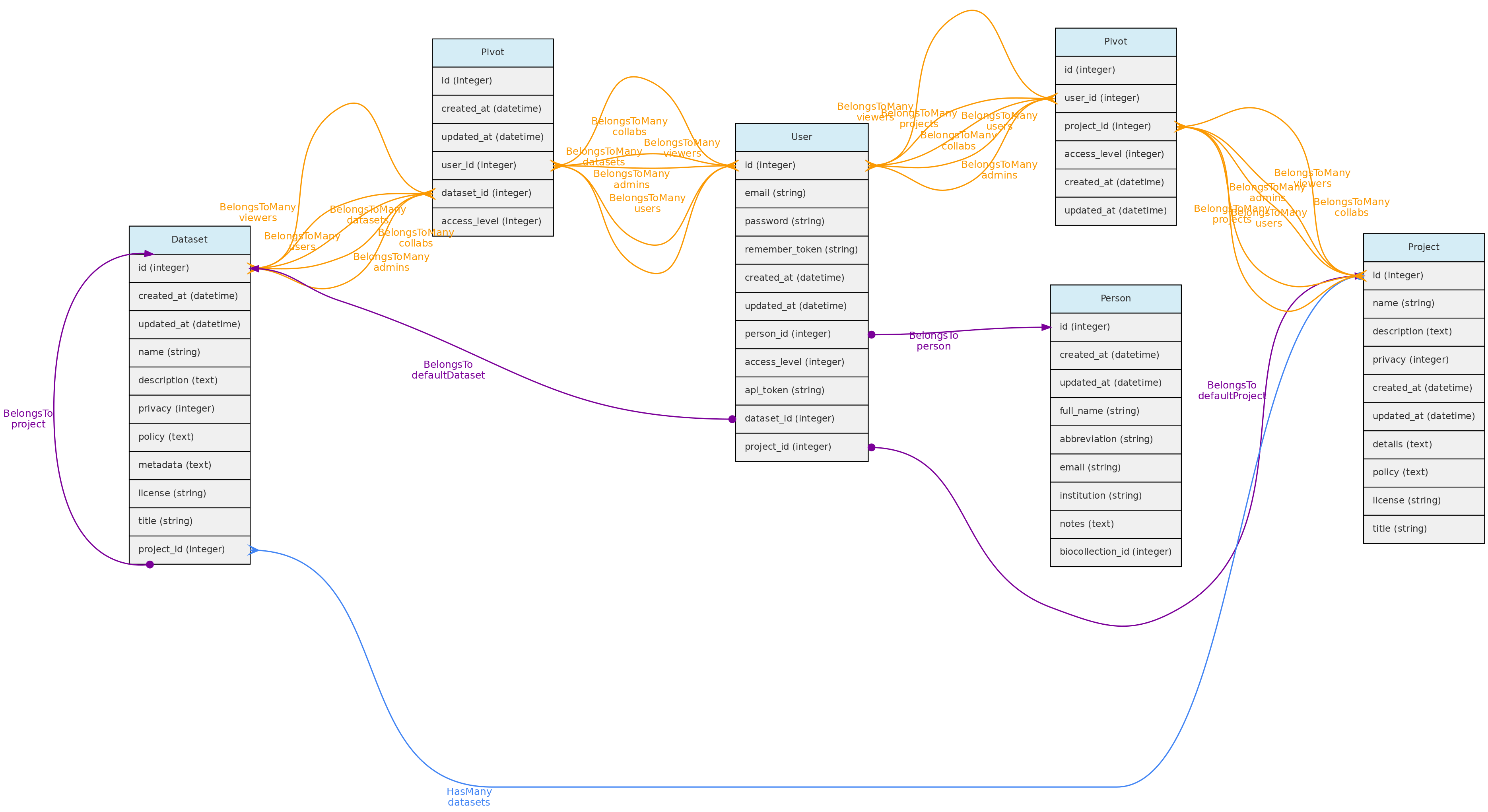
Each user is assigned to the registered user level when she or he registers in an OpenDataBio system. After that, a SuperAdmin may promote her/him to Full User or SuperAdmin. SuperAdmins also have the ability to edit other users and remove them from the database.
Every registered user is created along with a restricted Project and Dataset, which are referred to as her user Workspace. This allows users to import individual and voucher data before incorporating them into a larger project. [TO IMPLEMENT: export batches of objects from one project to another].
Data Access:users are created upon registration. Only administrators can update and delete user records.
User Jobs
The UserJob table is used to store temporarily background tasks, such as importing and exporting data. Any user is allowed to create a job; cancel their own jobs; list jobs that have not been deleted. The Job table contains the data used by the Laravel framework to interact with the Queue. The data from this table is deleted when the job runs successfully. The UserJob entity is used to keep this information, along with allowing for job logs, retrying failed jobs and canceling jobs that have not yet finished.
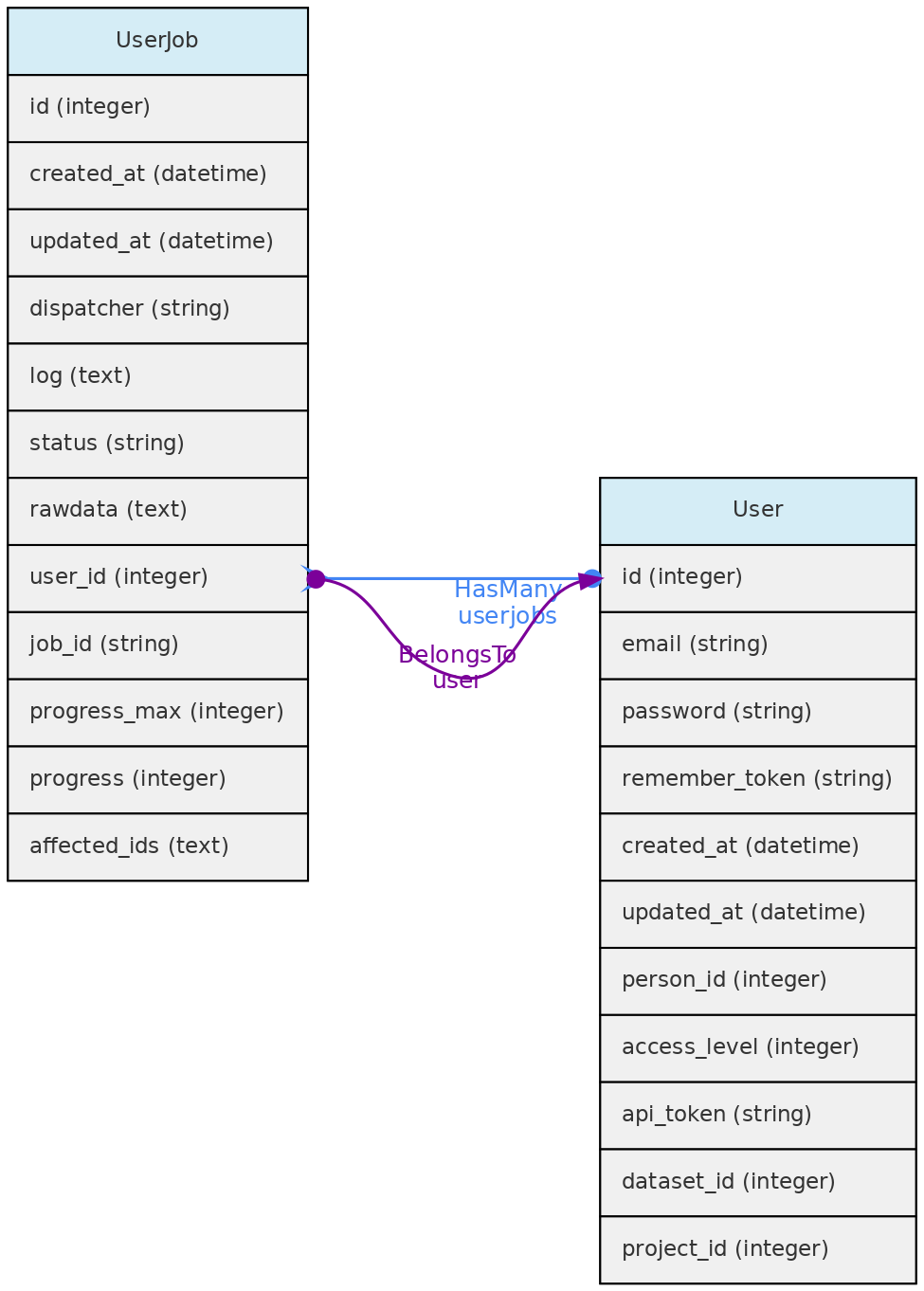
Data Access: Each registered user can see, edit and remove their own UserJobs.
4 - Auxiliary Objects
Libraries of common use like Persons and Bibliographic references and multilingual translations!
BibReference Model
The BibReference table contains basically BibTex formatted references stored in the bibtex column. You may easily import references into OpenDataBio by just specifying the doi, or simply uploading a bibtex record. These bibliographic references may be used to:
- Store references for Datasets - with the option of defining references for which citation is mandatory when using the dataset in publications; but all references that have used the dataset may be linked to the dataset; links are done with a Pivot table named
dataset_bibreference; - Store the references for Taxons:
- to specify the reference in which the Taxon name was described, currently mandatory in some Taxonomic journals like PhytoTaxa. This description reference is stored in the
bibreference_id of the Taxons table. - to register any reference to a Taxon name, which are then linked through a pivot table named
taxons_bibreference.
- Link a Measurement to a published source;
- Indicate the source of a Trait definition.
- Indicate mandatory citations for a Dataset, or link references using the data to a Dataset
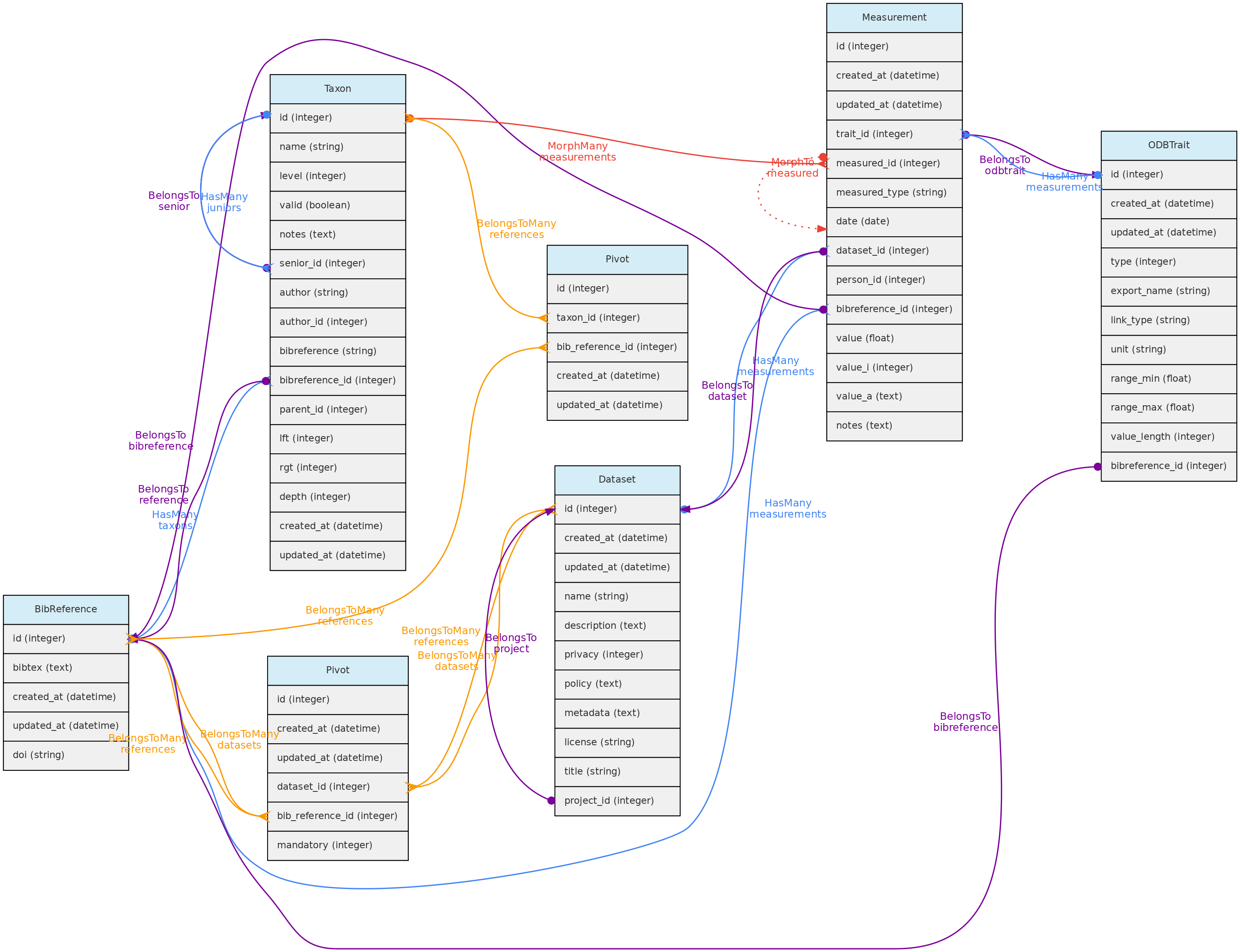 BibReference model and its relationships. Lines linking tables indicate the
BibReference model and its relationships. Lines linking tables indicate the methods implemented, with colors indicating different Eloquent relationships.
Bibreferences table
- The BibtexKey, authors and other relevant fields are extracted from the
bibtex column. - The Bibtexkey must be unique in the database, and a helper function is be provided to standardize it with format
<von? last name> <year> <first word of title>. The “von part” of the name is the “von”, “di”, “de la”, which are part of the last name for some authors. The first word of the title ignores common stop-words such as “a”, “the”, or “in”. - DOIs for a BibReference may be specified either in the relevant BibTex field or in a separate text input, and are stored in the
doi field when present. An external API finds the bibliographic record when a user informs the doi.
**Data access** [full users](/en/docs/concepts/data-access/#user) may register new references, edit references details and remove reference records that have no associated data. BibReferences have public access!
Identification Model
The Identification table represents the taxonomic identification of Individuals.
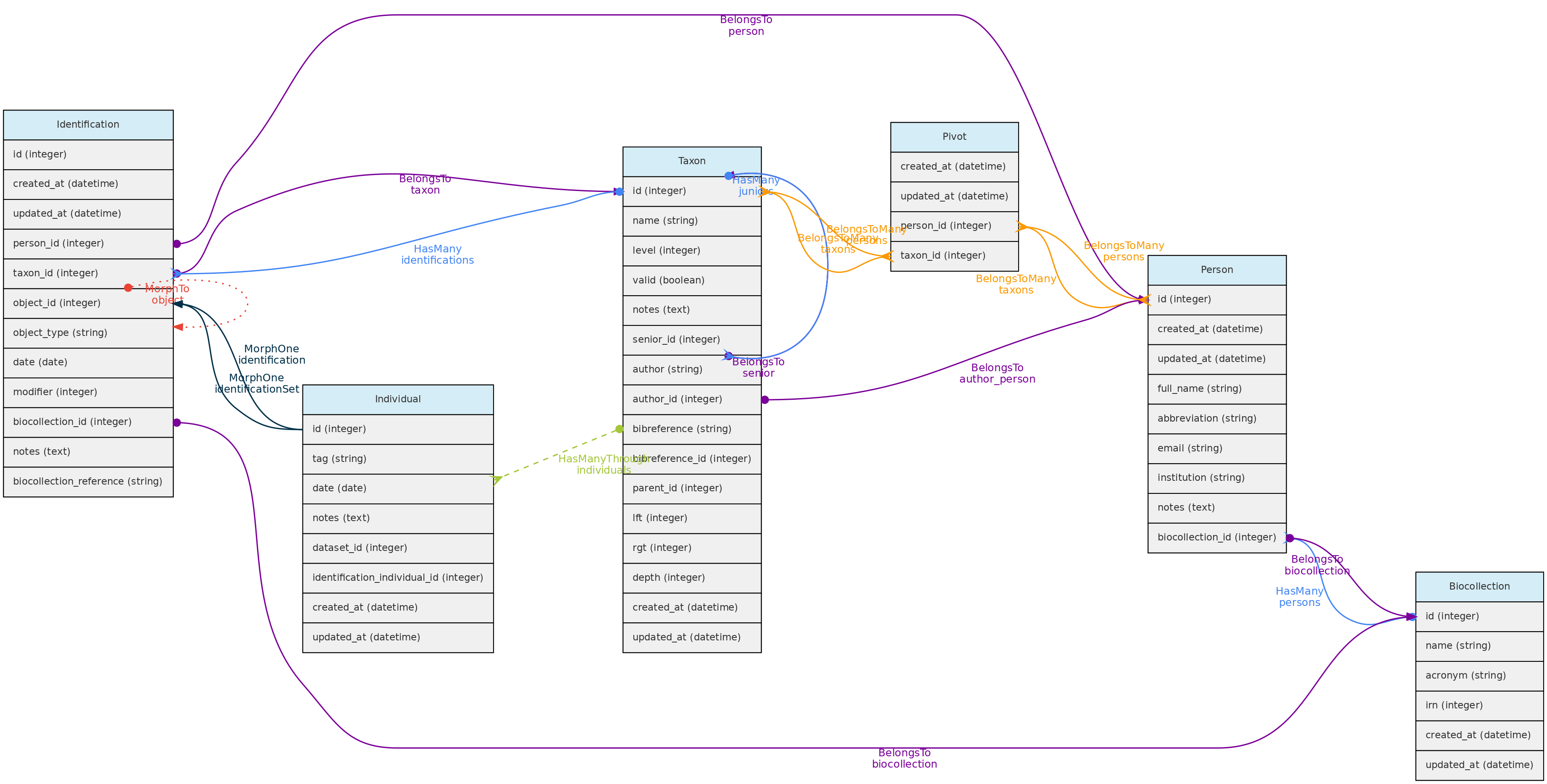 Identification model and its relationships. Lines linking tables indicate the
Identification model and its relationships. Lines linking tables indicate the methods implemented, with colors indicating different Laravel Eloquent relationships
Identifications table
- The Identification model includes several optional fields, but in addition to
taxon_id, person_id, the Person responsible for the identification, and the identification date are mandatory.
- The
date value may be an Incomplete Date, e.g. only the year or year+month may be recorded.
- The following fields are optional:
modifier - is a numeric code appending a taxonomic modifier to the name. Possible values ’s.s.’=1, ’s.l.’=2, ‘cf.’=3, ‘aff.’=4, ‘vel aff.’=5, defaults to 0 (none).notes - a text of choice, useful for adding comments to the identification.biocollection_id and biocollection_reference - these fields are to be used to indicate that the identification is based upon comparison to a voucher deposited in a Biological Collection and creates a link between the Individual identified and the BioCollection specimen from which the identification is based upon. biocollection_id stores the Biocollection id, and biocollection_reference the unique identifier of the specimen compared, i.e. would be the equivalent of the biocollection_number of the Voucher model, but this reference does not need to be from a voucher registered in the database.
- The relationship with the Individual model is defined by a polymorphic relationship using fields
object_type and object_id [This could be replaced by an ‘individual_id’ in the identification table. The polymorphic relation inherited from a previous development version, kept because the Identification model may be used in the future to link Identifications to Measurements]. - Changes in identifications are audited for tracking change history
Data access: identifications are attributes of Individuals and do not have independent access!
Person Model
The Person object stores persons names, which may or may not be a User directly involved with the database. It is used to store information about people that are:
* collectors of Vouchers, Individuals and MediaFiles
* taxonomic determinators or identifiers of individuals;
* measurer of Measurements;
* authors for unpublished Taxon names;
* taxonomic specialists - linked with Taxon model by a pivot table named person_taxon;
* dataset authors - defining authors for the dynamic publication of datasets;
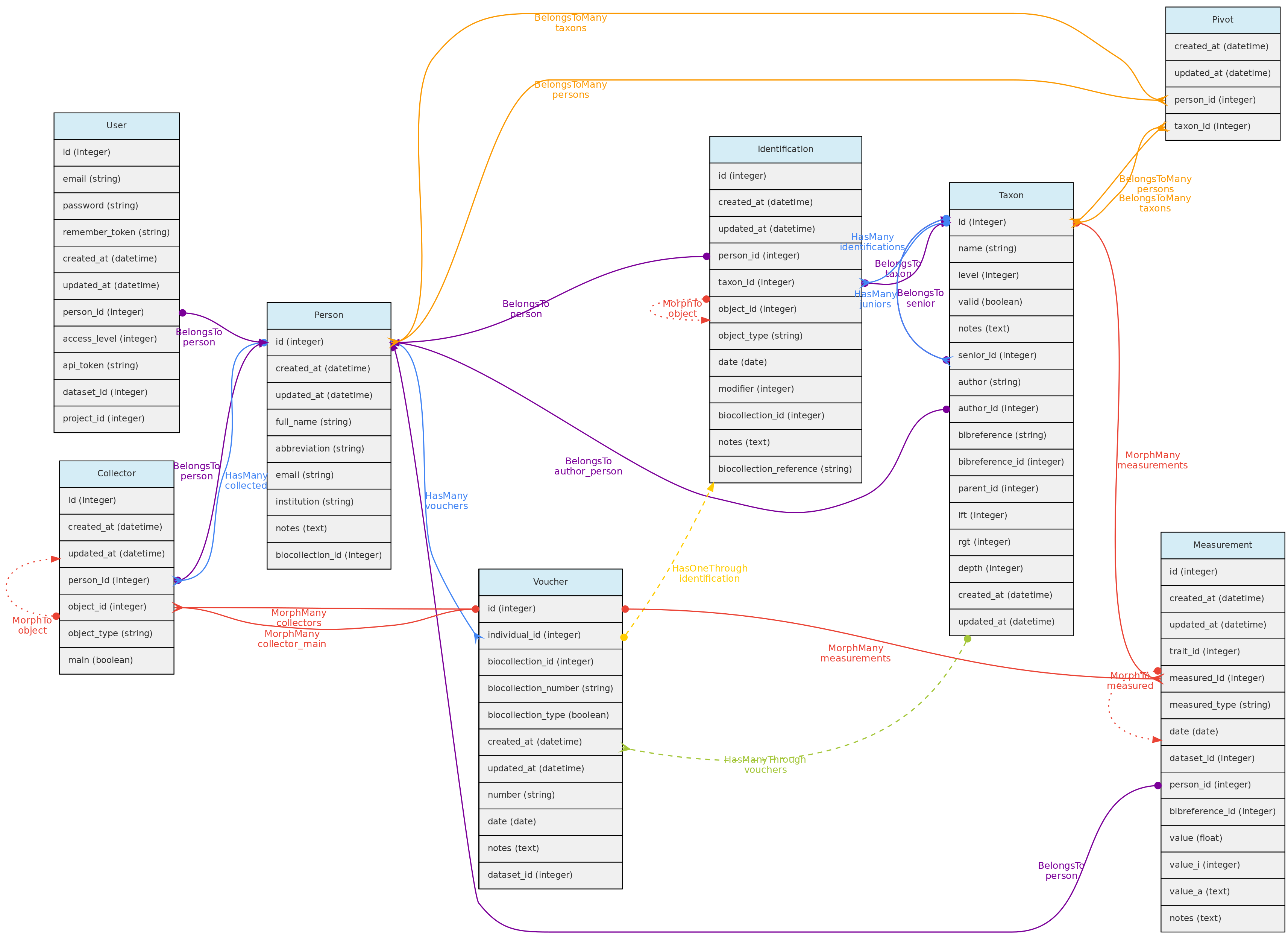 Person model and its relationships. Lines linking tables indicate the
Person model and its relationships. Lines linking tables indicate the methods implemented, with colors indicating different types of Laravel Eloquent methods, solid lines the direct and dashed the indirect relationships
Persons table
- mandatory columns are the person
full_name and abbreviation; - when registering a new person, the system suggests the name
abbreviation, but the user is free to change it to better adapt it to the usual abbreviation used by each person. The abbreviation must be unique in the database, duplicates are not allowed in the Persons table. Therefore, two persons with the exact same name must be differentiated somehow in the abbreviation column. - The
biocollection_id column of the Persons table is used to list to which Biocollection a person is associated, which may be used when the Person is also a taxonomic specialist. - Additionally, the
email and institution the person belongs to may also be informed. - Each user can be linked to a Person by the
person_id in the User table. This person is then used the ‘default’ person when the user is logged into the system.
**Data access** [full users](/en/docs/concepts/data-access/#user) may register new persons and edit the persons they have inserted and remove persons that have no associated data. Admins may edit any Person. Persons list have public access.
Media files are similar to measurements in that they might be associated with any core object. Media files may be images (jpeg, png, gif, tif), video or audio files and can be made freely accessible or placed in a Dataset with a defined access policy. A CreativeCommons.org license must be assigned to them. Media files may be tagged, i.e. you may define keywords to them, allowing to query them by Tags. For example, an individual image may be tagged with ‘flowers’ or ‘fruits’ to indicate what is in the image, or a tag that informs about image quality.
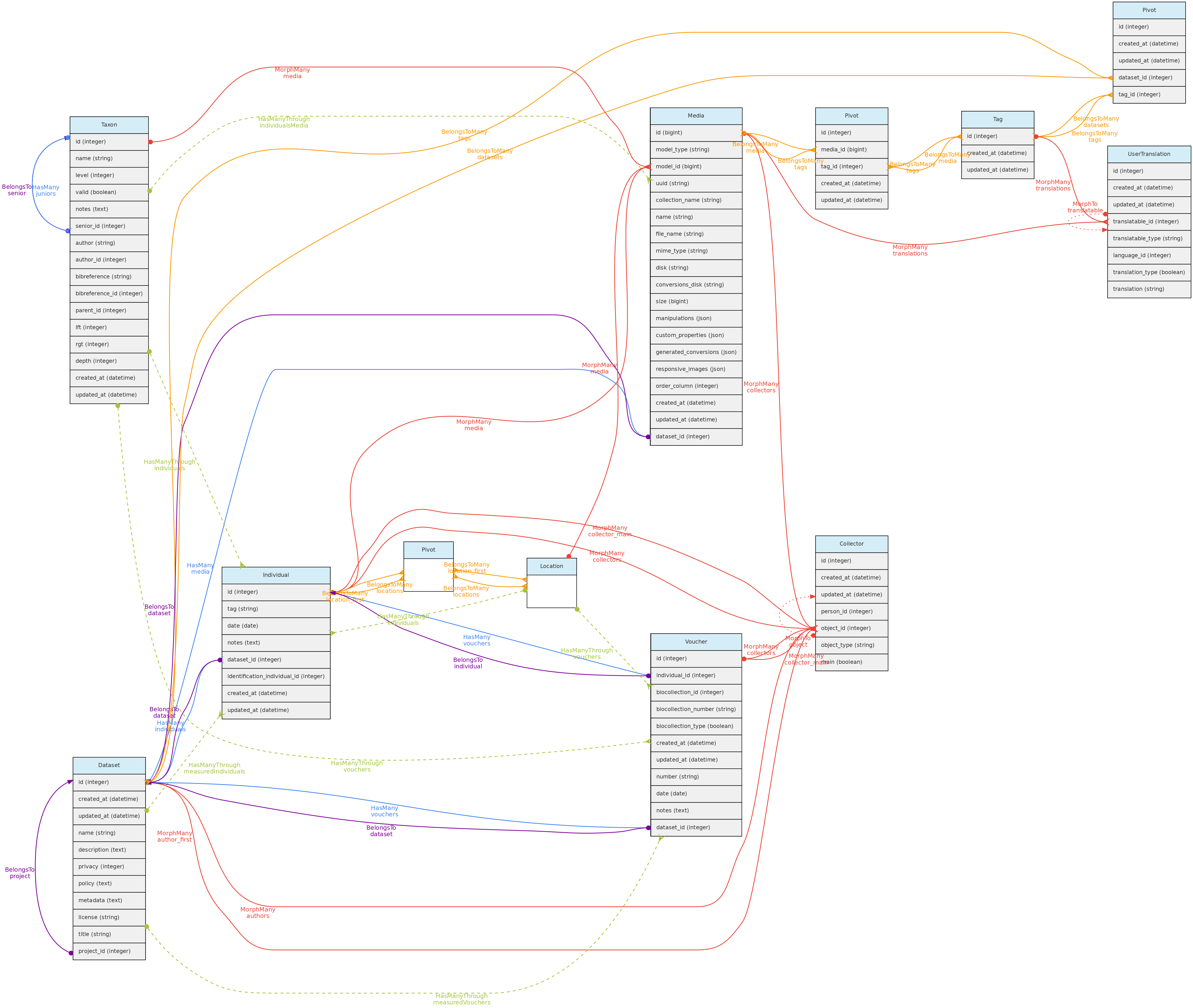
- Media files (image, video, audio) are linked to the Core-Objects through a polymorphic relationship defined by columns
model_id and model_type. - Multiple Persons may be associated with the Media for credits, these are linked with the Collectors table and its polymorphic relationship structure.
- A Media may have a
description in each language configured in the Language table, which will be stored in the user_translations table, which relates to the Tag model through a polymorphic relationship. Inputs for each language are shown in the web-interface forms. - Media files are not stored in the database, but in the server storage folder.
- It is possible to batch upload media files through the web interface, requiring also a file informing the objects to link the media with.
Data access full users may register media files and delete the ones they have inserted. If Media is in a Dataset, dataset admins may delete the media in addition to the user. Media files have public access, except when linked to a Dataset with access restrictions.
Tag Model
The Tag model allows users to define translatable keywords that may be used to flag Datasets, Projects or MediaFiles. The Tag model is linked with these objects through a pivot table for each, named dataset_tag, project_tag and media_tag, respectively.
A Tag may have name and description in each language configured in the Language table, which will be stored in the user_translations table, which relates to the Tag model through a polymorphic relationship. Inputs for each language are shown in the web-interface forms.
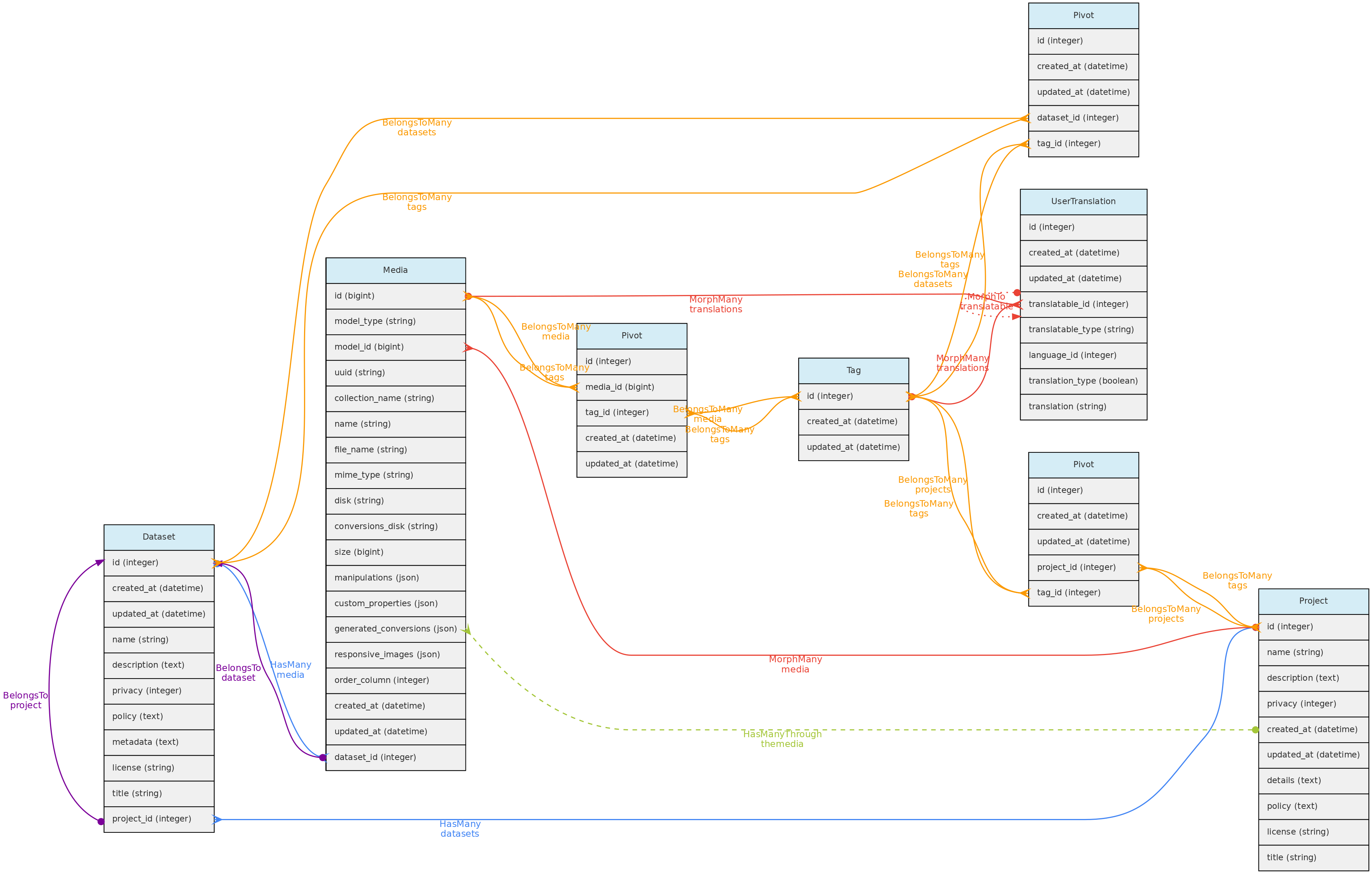
Data access full users may register tags, edit those they have inserted and delete those that have not been used. Tags have public access as they are just keywords to facilitate navigation.
User Translation Model
The UserTranslation model translates user data: Trait and Trait Categories names and descriptions, MediaFiles descriptions and Tags. The relations between these models are established by polymorphic relations using fields translatable_type and translatable_id. This model permits translations to any language listed in the Language table, which is currently only accessible for insertion and edition directly in the SQL database. Input forms in the web interface will be listed for registered Languages.
Vernacular - Popular Name
The Vernacular and VernacularCitation templates allow you to record popular names of organisms by relating them to Taxons and/or Individuals. Names must be unique in the Vernacular table, and each record can be linked to multiple Taxa and/or Individuals, depending on the information sources. Each record can also have one or more citations (citation text + BibReference + note).
Incomplete Dates
Dates for Vouchers, Individuals, Measurements and Identifications may be Incomplete, but at least year is mandatory in all cases. The date columns in the tables are of ‘date’ type, and incomplete dates are stored having 00 in the missing part: ‘2005-00-00’ when only year is known; ‘1988-08-00’ when only month is known.
Auditing
Modifications in database records are logged to the activity_log table. This table is generated by the package ActivityLog. The activities are shown in a ‘History’ link provided in the Show.view of the models.
- The package stores changes as json in the
properties field, which contains two elements: attributes and old, which are basically the new vs old values that have been changed. This structure must be respected. - Class ActivityFunctions contain custom functions to read the the properties Json record stored in the
activity_log table and finds the values to show in the History datatable; - Most changes are logged by the package as a ’trait’ called within the Class. These allow to automatically log most updates and are all configured to log only the fields that have changed, not entire records (option
dirty). Also record creation are not logged as activity, only changes. - Some changes, like Individual and Vouchers collectors and identifications are manually logged, as they involve related tables and logging is specified in the Controller files;
- Logging contain a
log_name field that groups log types, and is used to distinguish types of activity and useful to search the History Datatable; - Two special logging are also done:
- Any Dataset download is logged, so administrators may track who and when the dataset was downloaded;
- Any Dataset request is also logged for the same reason
The clean-command of the package SHOULD NOT be used during production, otherwise will just erase all logged changes. If run, will erase the logs older than the time specified in the /config/activitylog.php file.
The ActivityLog table has the following structure:
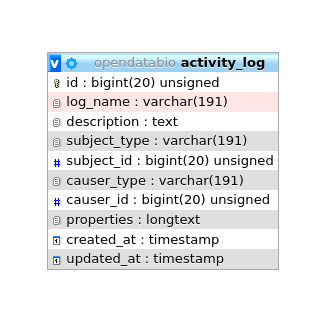
causer_type and causer_id will be the User that made the changesubject_type and subject_id will the model and record changedlog_name - to group logs together and permits queriesdescription - somewhat redundant with log_name in a OpenDataBio context.properties - stores the changes, for example, and identification change will have a log like:
{
"attributes":
{
"person_id":"2",
"taxon_id":"1424",
"modifier":"2",
"biocollection_id":"1",
"biocollection_reference":"1234",
"notes":"A new fake note has been inserted",
"date":"2020-02-08"},
"old":{
"person_id":674,
"taxon_id":1413,
"date":"1995-00-00",
"modifier":0,
"biocollection_id":null,
"notes":null,
"biocollection_reference":null
}
}

 This figure shows the relationships of the
This figure shows the relationships of the  The same tables related with the Location model with the direct and non-polymoprhic relationships indicated.
The same tables related with the Location model with the direct and non-polymoprhic relationships indicated. This figure shows the Individual Model and the models it relates to, except the Measurement and Location models, as their relationships with Individuals is shown elsewhere in this page. Lines linking models indicate the
This figure shows the Individual Model and the models it relates to, except the Measurement and Location models, as their relationships with Individuals is shown elsewhere in this page. Lines linking models indicate the  The Individual model direct and non-polymoprhic relationships.
The Individual model direct and non-polymoprhic relationships. Taxon model and its relationships. Lines linking tables indicate the
Taxon model and its relationships. Lines linking tables indicate the  Voucher model and its relationships. Lines linking tables indicate the
Voucher model and its relationships. Lines linking tables indicate the 







 BibReference model and its relationships. Lines linking tables indicate the
BibReference model and its relationships. Lines linking tables indicate the  Identification model and its relationships. Lines linking tables indicate the
Identification model and its relationships. Lines linking tables indicate the  Person model and its relationships. Lines linking tables indicate the
Person model and its relationships. Lines linking tables indicate the 


