Getting data with OpenDataBio-R
7 minute read
The Opendatabio-R package was created to allow users to interact with an OpenDataBio server, to both obtain (GET) data or to import (POST) data into the database. This tutorial is a basic example of how to get data.
Set up the connection
- Set up the connection to the OpenDataBio server using the
odb_config()function. The most important parameters for this function arebase_url, which should point to the API url for your OpenDataBio server, andtoken, which is the access token used to authenticate your user. - The
tokenis only need to get data from datasets that have one of the restricted access policies. Data from datasets of public access can be extracted without the token specification. - Your token is avaliable in your profile in the web interface
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
More advanced configuration involves setting a specific API version, a custom User Agent, or other HTTP headers, but this is not covered here.
Test your connection
The function odb_test() may be used to check if the connection was successful, and whether
your user was correctly identified:
odb_test(cfg)
#will output
Host: https://opendb.inpa.gov.br/api/v0
Versions: server 0.9.1-alpha1 api v0
$message
[1] "Success!"
$user
[1] "admin@example.org"
As an alternative, you can specify these parameters as systems variables. Before starting R, set this up on your shell (or add this to the end of your .bashrc file):
export ODB_TOKEN="YourToken"
export ODB_BASE_URL="https://opendb.inpa.gov.br/api"
export ODB_API_VERSION="v0"
GET Data
See the GET API Quick Reference for a complete list of endpoints and request parameters. Also see the generic parameters, especially
save_jobwhich is important for downloading large datasets.
For publicly accessible data the token is optional. Below are some examples. Follow a similar reasoning to use the other endpoints. See the R package help for all available odb_get_{endpoint} functions.
Getting Taxon names
See GET API Taxon Endpoint request parameters and a list of response fields.
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
#get id for a taxon
mag.id = odb_get_taxons(params=list(name='Magnoliidae',fields='id,name'),odb_cfg = cfg)
#use this id to get all descendants of this taxon
odb_taxons = odb_get_taxons(params=list(root=mag.id$id,fields='id,scientificName,taxonRank,parent_id,parentName'),odb_cfg = cfg)
head(odb_taxons)
If the server used the seed data provided and the default language is portuguese, the result will be:
id scientificName taxonRank parent_id parentName
1 25 Magnoliidae Clado 20 Angiosperms
2 43 Canellales Ordem 25 Magnoliidae
3 62 Laurales Ordem 25 Magnoliidae
4 65 Magnoliales Ordem 25 Magnoliidae
5 74 Piperales Ordem 25 Magnoliidae
6 93 Chloranthales Ordem 25 Magnoliidae
Getting Locations
See GET API Location Endpoint request parameters and a list of response fields. See also the POST Locations-Validation Endpoint if you have latitude and longitude and wants to validade the geometries and find where the point falls.
Get some fields listing all Conservation Units (adm_level==99) registered in the server:
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
odblocais = odb_get_locations(params = list(fields='id,name,parent_id,parentName',adm_level=99),odb_cfg = cfg)
head(odblocais)
If the server used the seed data provided and the default language is portuguese, the result will be:
id name
1 5628 Estação Ecológica Mico-Leão-Preto
2 5698 Área de Relevante Interesse Ecológico Ilha do Ameixal
3 5700 Área de Relevante Interesse Ecológico da Mata de Santa Genebra
4 5703 Área de Relevante Interesse Ecológico Buriti de Vassununga
5 5707 Reserva Extrativista do Mandira
6 5728 Floresta Nacional de Ipanema
parent_id parentName
1 6 São Paulo
2 6 São Paulo
3 6 São Paulo
4 6 São Paulo
5 6 São Paulo
6 6 São Paulo
Locations as spatial objects in R
To obtain a spatial object in R, use the sf package. The example below plots a plot and its subplots and also exports the locations as both, kml and shapefile.
library(sf)
library(opendatabio)
#download all plot locations
cfg <- odb_config(base_url = "https://opendb.inpa.gov.br/api")
#get a large main plot
parcela = odb_get_locations(params = list(fields='all',name='Parcela 25ha'), odb_cfg = cfg)
parcela$type = 'main plot'
#get subplots
subplots = odb_get_locations(params = list(fields='all',location_root=parcela$id), odb_cfg = cfg)
subplots = subplots[subplots$adm_level==100 & subplots$id!=parcela$id,]
subplots$type = 'sub plot'
#convert footprintWKT to sf geometries
geoms <- st_as_sfc(parcela$footprintWKT, crs = 4326)
parcela_sf <- st_sf(parcela, geometry = geoms)
geoms <- st_as_sfc(subplots$footprintWKT, crs = 4326)
subplots_sf <- st_sf(subplots, geometry = geoms)
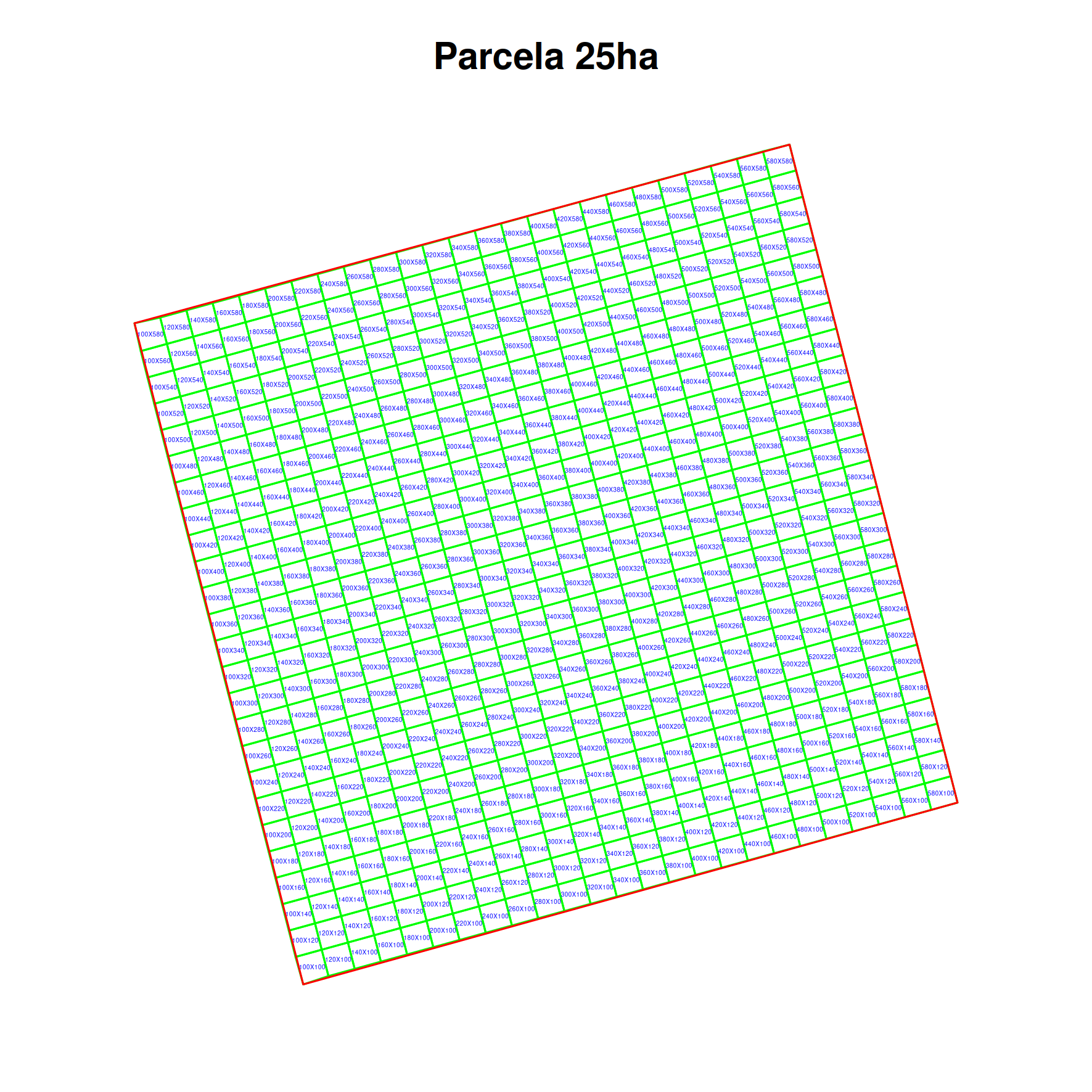
#print a figure
png("plots_with_subplots.png", width = 15, height = 15,units='cm',res=300)
par(mar=c(2,2,3,2))
plot(st_geometry(subplots_sf),border='green',main = parcela$locationName)
plot(st_geometry(parcela_sf),border='red',add=T)
labs = gsub("Quadrat ","",subplots_sf$locationName)
text(
st_coordinates(st_centroid(subplots_sf)),
labels = labs,
cex = 0.2, col = "blue"
)
dev.off()
#save as kml
locais = rbind(parcela_sf,subplots_sf)
locais$name <- locais$locationName
cols_to_include <- setdiff(names(locais), c("footprintWKT",'locationName'))
locais_kml <- locais[, c("name", cols_to_include[cols_to_include != "name"])]
st_write(locais_kml, "plots_and_subplots.kml", layer=parcela$locationName, driver = "KML", delete_dsn = TRUE)
#save as shapefile
st_write(locais_kml, "plots_and_subplots.shp", layer=parcela$locationName, delete_layer = TRUE)
Figure generated:
Validating point geometries
See the POST Locations-Validation Endpoint.
#conect to database
library(opendatabio)
base_url="http://localhost/opendatabio/api"
token ="your token is mandatory in this case"
cfg = odb_config(base_url=base_url, token = token)
odb_test(cfg)
#fake data
dados = data.frame(
latitude = sample(seq(-2,2,by=0.00001),10),
longitude = sample(seq(-60,-59,by=0.00001),10)
)
#submit job for validation
jb = odb_validate_locations(dados,odb_cfg = cfg)
#monitor job execution
odb_get_jobs(params=list(id=jb$id),odb_cfg = cfg)
#get results
dadosValidados = odb_get_jobs(params=list(id=jb$id,get_file=T),odb_cfg = cfg)
head(dados)
latitude longitude
1 0.12975 -59.65745
2 1.77469 -59.77757
3 -0.89154 -59.80179
4 -1.25632 -59.87084
5 0.77085 -59.22740
6 -0.74237 -59.64591
head(dadosValidados)
latitude longitude withinLocationName withinLocationParent withinLocationCountry withinLocationHigherGeography withinLocationType
1 0.12975 -59.65745 Trombetas/Mapuera Brasil Brazil Brasil > Trombetas/Mapuera Território Indígena
2 0.12975 -59.65745 Bioma Amazônia Brasil Brazil Brasil > Bioma Amazônia Ambiental
3 0.12975 -59.65745 Amazonia World Amazonia Ambiental
4 0.12975 -59.65745 Urucará Amazonas Brazil Brasil > Amazonas > Urucará Município
5 1.77469 -59.77757 Jacamim Roraima Brazil Brasil > Roraima > Jacamim Território Indígena
6 1.77469 -59.77757 Bioma Amazônia Brasil Brazil Brasil > Bioma Amazônia Ambiental
withinLocationID withinLocationTypeAdmLevel searchObs
1 6393 98 NA
2 6583 97 NA
3 16597 97 NA
4 1570 8 NA
5 6121 98 NA
6 6583 97 NA
Getting Individual Data
See GET API Individual Endpoint for the full list of search parameters and response fields.
library(opendatabio)
base_url = "https://opendb.inpa.gov.br/api"
token = "YOUR TOKEN HERE"
# Set the connection configuration
cfg = odb_config(base_url = base_url, token = token)
# DIRECT DOWNLOAD – if you want to download a small amount of data
inds = odb_get_individuals(params = list(limit = 100), odb_cfg = cfg)
# PREPARE FILE ON SERVER – if your query will return a large number of records
# Download all records you have access to or public ones
# Save the process, since the result is likely to be large
jobid = odb_get_individuals(params = list(save_job = TRUE), odb_cfg = cfg)
# Check the status of the job
odb_get_jobs(params = list(id = jobid$job_id), odb_cfg = cfg)
# When it finishes, get the data here (or download the file via the web interface)
all_inds = odb_get_jobs(params = list(id = jobid$job_id), odb_cfg = cfg)
# FETCHING SPECIFIC DATA
# All individuals identified as taxon X
params = list(taxon = "Licaria cannela tenuicarpa")
licarias = odb_get_individuals(params = params, odb_cfg = cfg)
# All individuals identified as taxon X or its descendants
params = list(taxon_root = "Licaria")
licarias = odb_get_individuals(params = params, odb_cfg = cfg)
# All individuals from dataset X
params = list(dataset = "MyDataset name or id")
inds = odb_get_individuals(params = params, odb_cfg = cfg)
# Or use save_job above if the dataset is large
# You can view the list of available datasets
datasets = odb_get_datasets(odb_cfg = cfg)
Getting Measurements
See GET API Measurement Endpoint for the complete list of query parameter options and response fields.
Use the odb_get_measurements function.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token="YOUR TOKEN HERE"
#establishes the connection configuration
cfg = odb_config(base_url=base_url, token = token)
#100 first measurements of the dataset X with id=10
measurements = odb_get_measurements(params=list(dataset=10,limit=100),odb_cfg=cfg)
#100 first measurements of the dataset X with id=10 for the variable whose export_name is treeDbh
measurements = odb_get_measurements(params=list(trait="treeDbh",dataset=10,limit=100),odb_cfg=cfg)
#Measurements of the dataset X with id=10 for the variable whose export_name is treeDbh
#only for Lauraceae
measurements = odb_get_measurements(params=list(trait="treeDbh",dataset=10,taxon_root="Lauraceae"),odb_cfg=cfg)
#linking data of individuals measurements
laurels = odb_get_individuals(params=list(dataset=10,taxon_root="Lauraceae"),odb_cfg=cfg)
filter = grep("Individu",measurements$measured_type) #optional, depends on what is in measurements
g = match(measurements$measured_id[filter],laurels$id)
measurements$location = NA
measurements$location[filter] = laurels$locationName[g]
Getting Media
See GET API Media Endpoint for the complete list of query parameter options and response fields.
Use the odb_get_media function from the R package.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token="YOUR TOKEN HERE"
#set the connection configuration
cfg = odb_config(base_url=base_url, token = token)
#the first 50 media files of a dataset that has images
imgs = odb_get_media(params=list(dataset=97,limit=50),odb_cfg=cfg)
#see this metadata
head(imgs)
#from this metadata, download the media files
#create a function for this:
getImagesByURL <- function(url,downloadFolder='img') {
dir.create(downloadFolder,showWarnings = F)
fn = strsplit(url,"\\/")[[1]]
fn = fn[length(fn)]
nname = paste(downloadFolder,fn,sep="/")
img = httr::GET(url=url)
writeBin(httr::content(img, "raw"), nname)
}
#use the function to download images to a folder
sapply(imgs$file_url,getImagesByURL,downloadFolder='testeImgsFromOdb')
Getting Voucher Data
See GET API Voucher Endpoint for the full list of search parameter options and response fields.
Follow the example above, but use the odb_get_vouchers function.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token="YOUR TOKEN HERE"
#establishes the connection configuration
cfg = odb_config(base_url=base_url, token = token)
#first 100 vouchers registered in a biocollection
vouchers = odb_get_vouchers(params=list(biocollection="INPA",limit=100),odb_cfg=cfg)
#vouchers in location x (id, or name, as registered in the database)
vouchers = odb_get_vouchers(params=list(location="Reserva Florestal Adolpho Ducke, Parcela PDBFF-100ha",limit=100),odb_cfg=cfg)