Import data with R
Import data using the OpenDataBio R client
The Opendatabio-R package was created to allow users to interact with an OpenDataBio server, to both obtain (GET) data or to import (POST) data into the database. This tutorial is a basic example of how to import data.
Set up the connection
- Set up the connection to the OpenDataBio server using the
odb_config() function. The most important parameters for
this function are base_url, which should point to the API url for your OpenDataBio server, and
token, which is the access token used to authenticate your user. - The
token is mandatory for importing data. - Your token is avaliable in your profile in the web interface
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
#create a config object
cfg = odb_config(base_url=base_url, token = token)
#test connection
odb_test(cfg)
Importing data (POST API)
Check the API Quick-Reference for a full list of POST endpoints and link to details.
OpenDataBio-R import functions
All import functions have the same signature: the first argument is a data.frame with data to be imported, and the second parameter is a configuration object generated by odb_config.
When writing an import request, check the POST API docs in order to understand which columns can be declared in the data.frame.
All import functions return a job id, which can be used to check if the job is still running, if it ended with success or if it encountered an error. This job id can be used in the functions odb_get_jobs(), odb_get_affected_ids() and odb_get_log(), to find details about the job, which (if any) were the IDs of the successfully imported objects, and the full log of the job. You may also see the log in your user jobs list in the web interface.
Attention
Order is Important - correctly importing data may depend on already registered records. For example, importing an Individual with a Taxon identity requires the taxon name registered in the database, so you would first validate your taxon list using the GET API and then import the records for the Individuals.
You may also Validate geographic coordinates, detecting locations registered in an OpenDataBio database for point geometries.
Working with dates and incomplete dates
For Individuals, Vouchers and Identifications you may use incomplete dates.
The date format used in OpenDataBio is YYY-MM-DD (year - month - day), so a valid entry would be 2018-05-28.
Particularly in historical data, the exact day (or month) may not be known, so you can substitute this fields with NA: ‘1979-05-NA’ means “an unknown day, in May 1979”, and ‘1979-NA-NA’ means “unknown day and month, 1979”. You may not add a date for which you have only the day, but can if you have only the month if is actually meaningful in some way.
1 - Import Locations
Import locations using the OpenDataBio R client
OpenDataBio is distributed with a seed location dataset for Brazil, which includes state, municipality, federal conservation units, indigenous lands and the major biomes.
Working with spatial data is a very delicate area, so we have attempted to make the workflow for inserting locations as easy as possible.
If you want to upload administrative boundaries for a country, you may also just download a geojson file from OSM-Boundaries and upload it through the web interface directly. Or use the GADM repository exemplified below.
Importation is straightforward, but the main issues to keep in mind:
- OpenDataBio stores the geometries of locations using Well-known text (WKT) representation.
- Locations are hierarchical, so a location SHOULD lie completely within its parent location. The importation method will try to detect the parent locations based on its geometry. So you do not need to inform a parent. However, sometimes the parent and child locations share a border or have minor differences that prevent to be detected. Therefore, if this importation fail to place the location where you expected, you may update or import informing the correct parent. When you inform the parent a second check will be performed adding a buffer to the parent location, and should solve the issue.
- Country borders can be imported without parent detection or definition, and marine records may be linked to a parent even if they are not contained by the parent polygon. This requires a specific field specification and should be used only in such cases as it is a possible source of misplacement, but give such flexibility)
- Standardize the geometry to a common projection of use in the system. Strongly recommended to use EPSG:4326 WGS84., for standardization;
- Consider, uploading your political administrative polygons before adding specific POINT, PLOTS or TRANSECTS;
- Conservation Units, Indigenous Territories and Environmental layers may be added as locations and will be treated as special case as some of these locations span different administrative locations. So a POINT, PLOT or TRANSECT location may belong to a UC, a TI and many Environmental layers if these are stored in the database. These related locations, like the political parent, are auto-detected from the location geometry.
Check the POST Locations API docs in order to understand which columns can be declared when importing locations.
Adm_level defines the location type
The administrative level (adm_level) of a location is a number:
- 2 for country; 3 to 10 as other as ‘administrative areas’, following OpenStreeMap convention to facilitate external data importation and local translations (TO BE IMPLEMENTED). So, for Brazil, codes are (States=4, Municipalities=8);
- 999 for ‘POINT’ locations like GPS waypoints;
- 101 for transects
- 100 is the code for plots and subplots;
- 99 is the code for Conservation Units
- 98 for Indigenous Territories
- 97 for Environmental polygons (e.g. Floresta Ombrofila Densa, or Bioma Amazônia)
Importing spatial polygons
GADM Administrative boundaries
Administrative boundaries may also be imported without leaving R, getting data from GDAM and using the odb_import* functions
library(raster)
library(opendatabio)
#download GADM administrative areas for a country
#get country codes
crtcodes = getData('ISO3')
bra = crtcodes[crtcodes$NAME%in%"Brazil",]
#define a path where to save the downloaded spatial data
path = "GADMS"
dir.create(path,showWarnings = F)
#the number of admin_levels in each country varies
#get all levels that exists into your computer
runit =T
level = 0
while(runit) {
ocrt <- try(getData('GADM', country=bra, level=level,path=path),silent=T)
if (class(ocrt)=="try-error") {
runit = FALSE
}
level = level+1
}
#read downloaded data and format to odb
files = list.files(path, full.name=T)
locations.to.odb = NULL
for(f in 1:length(files)) {
ocrt <- readRDS(files[f])
#class(ocrt)
#convert the SpatialPolygonsDataFrame to OpenDataBio format
ocrt.odb = opendatabio:::sp_to_df(ocrt) #only for GADM data
locations.to.odb = rbind(locations.to.odb,ocrt.odb)
}
#see without geometry
head(locations.to.odb[,-ncol(locations.to.odb)])
#you may add a note to location
locations.to.odb$notes = paste("Source gdam.org via raster::get_data()",Sys.Date())
#adjust the adm_level to fit the OpenStreeMap categories
ff = as.factor(locations.to.odb$adm_level)
(lv = levels(ff))
levels(ff) = c(2,4,8,9)
locations.to.odb$adm_level = as.vector(ff)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
odb_import_locations(data=locations.to.odb,odb_cfg=cfg)
#ATTENTION: you may want to check for uniqueness of name+parent rather than just name, as name+parent are unique for locations. You may not save two locations with the same name within the same parent.
A ShapeFile example
library(rgdal)
#read your shape file
path = 'mymaps'
file = 'myshapefile.shp'
layer = gsub(".shp","",file,ignore.case=TRUE)
data = readOGR(dsn=path, layer= layer)
#you may reproject the geometry to standard of your system if needed
data = spTransform(data,CRS=CRS("+proj=longlat +datum=WGS84"))
#convert polygons to WKT geometry representation
library(rgeos)
geom = rgeos::writeWKT(data,byid=TRUE)
#prep import
names = data@data$name #or the column name of the data
shape.to.odb = data.frame(name=names,geom=geom,stringsAsFactors = F)
#need to add the admin_level of these locations
shape.to.odb$admin_level = 2
#and may add parent and note if your want
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
odb_import_locations(data=shape.to.odb,odb_cfg=cfg)
Converting data from KML
#read file as SpatialPolygonDataFrame
file = "myfile.kml"
file.exists(file)
mykml = readOGR(file)
geom = rgeos::writeWKT(mykml,byid=TRUE)
#prep import
names = mykml@data$name #or the column name of the data
to.odb = data.frame(name=names,geom=geom,stringsAsFactors = F)
#need to add the admin_level of these locations
to.odb$admin_level = 2
#and may add parent or any other valid field
#import
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
odb_import_locations(data=to.odb,odb_cfg=cfg)
Import Plots and Subplots
Plots and Transects are special cases within OpenDataBio:
- They may be defined with a Polygon or LineString geometry, respectively;
- Or they may be registered only as POINT locations. In this case OpenDataBio will create the polygon or linestring geometry for you;
- Dimensions (x and y) are stored in meters
- SubPlots are plot locations having a plot location as a parent, and must also have cartesian positions (startX, startY) within the parent location in addition to dimensions. Cartesian position refer to X and Y positions within parent plot and hence MUST be smaller than parent X and Y. And the same is true for Individuals within plots or subplots when they have their own X and Y cartesian coordinates.
- SubPlot is the only location that may be registered without a geographical coordinate or geometry, which will be calculated from the parent plot geometry using the startx and starty values.
Plot and subplot example 01
You need at least a single point geographical coordinate for a location of type PLOT. Geometry (or lat and long) cannot be empty.
#geometry of a plot in Manaus
southWestCorner = c(-59.987747, -3.095764)
northWestCorner = c(-59.987747, -3.094822)
northEastCorner = c(-59.986835,-3.094822)
southEastCorner = c(-59.986835,-3.095764)
geom = rbind(southWestCorner,northWestCorner,northEastCorner,southEastCorner)
library(sp)
geom = Polygon(geom)
geom = Polygons(list(geom), ID = 1)
geom = SpatialPolygons(list(geom))
library(rgeos)
geom = writeWKT(geom)
to.odb = data.frame(name='A 1ha example plot',x=100,y=100,notes='a fake plot',geom=geom, adm_level = 100,stringsAsFactors=F)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
odb_import_locations(data=to.odb,odb_cfg=cfg)
Wait a few seconds, and then import subplots to this plot.
#import 20x20m subplots to the plot above without indicating a geometry.
#SubPlot is the only location type that does not require the specification of a geometry or coordinates,
#but it requires specification of startx and starty relative position coordinates within parent plot
#OpenDataBio will use subplot position values to calculate its geographical coordinates based on parent geometry
(parent = odb_get_locations(params = list(name='A 1ha example plot',fields='id,name',adm_level=100),odb_cfg = cfg))
sub1 = data.frame(name='sub plot 40x40',parent=parent$id,x=20,y=20,adm_level=100,startx=40,starty=40,stringsAsFactors=F)
sub2 = data.frame(name='sub plot 0x0',parent=parent$id,x=20,y=20,adm_level=100,startx=0,starty=0,stringsAsFactors=F)
sub3 = data.frame(name='sub plot 80x80',parent=parent$id,x=20,y=20,adm_level=100,startx=80,starty=80,stringsAsFactors=F)
dt = rbind(sub1,sub2,sub3)
#import
odb_import_locations(data=dt,odb_cfg=cfg)
Screen captures of imported plots
Below screen captures for the locations imported with the code above
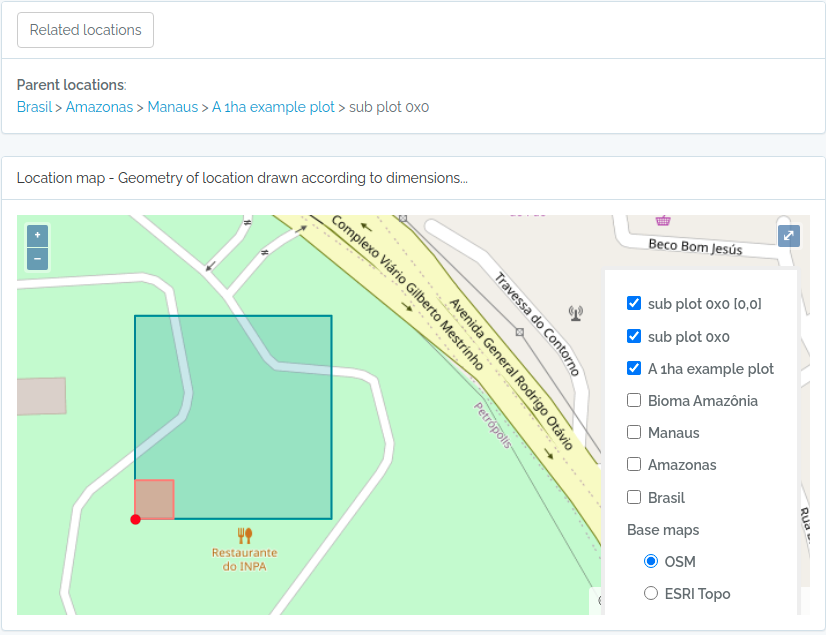
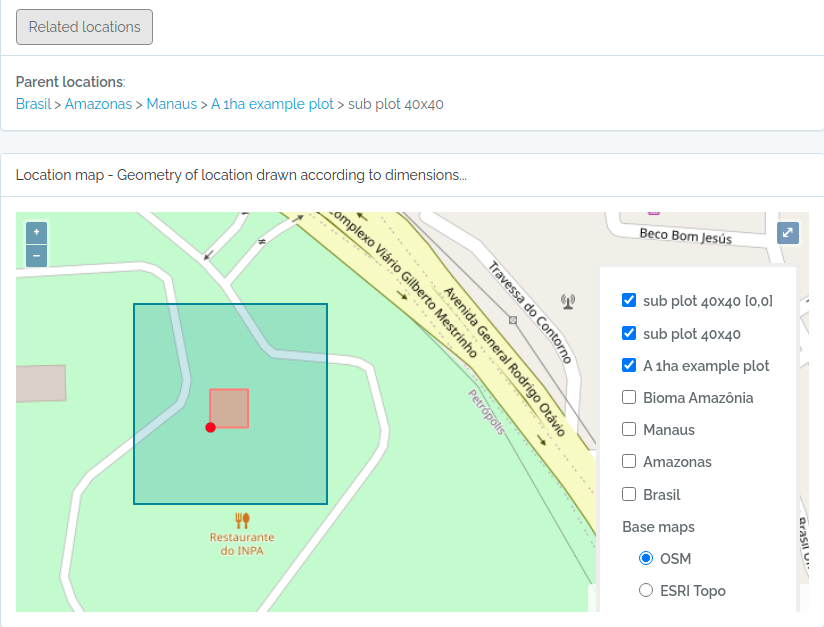
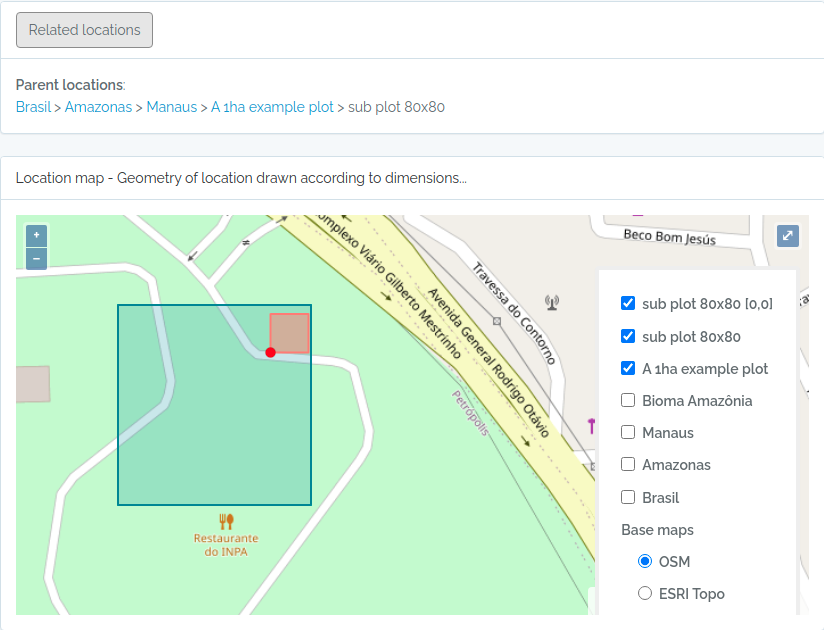
Plot and subplot example 02
Import a plot and subplots having only:
- a single point coordinate
- an azimuth or angle of the plot direction
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
#the plot
geom = "POINT(-59.973841 -2.929822)"
to.odb = data.frame(name='Example Point PLOT',x=100, y=100, azimuth=45,notes='OpenDataBio point plot example',geom=geom, adm_level = 100,stringsAsFactors=F)
odb_import_locations(data=to.odb,odb_cfg=cfg)
#define 20x20 subplots cartesian coordinates
x = seq(0,80,by=20)
xx = rep(x,length(x))
yy = rep(x,each=length(x))
names = paste(xx,yy,sep="x")
#import these subplots without having a geometry, but specifying the parent plot location
parent = odb_get_locations(params = list(name='Example Point PLOT',adm_level=100),odb_cfg = cfg)
to.odb = data.frame(name=names,startx=xx,starty=yy,x=20,y=20,notes="OpenDataBio 20x20 subplots example",adm_level=100,parent=parent$id)
odb_import_locations(data=to.odb,odb_cfg=cfg)
#get the imported plot locations and plot them using the root parameter
locais = odb_get_locations(params=list(root=parent$id),odb_cfg = cfg)
locais[,c('id','locationName','parentName')]
colnames(locais)
for(i in 1:nrow(locais)) {
geom = readWKT(locais$footprintWKT[i])
if (i==1) {
plot(geom,main=locais$locationName[i],cex.main=0.8,col='yellow')
axis(side=1,cex.axis=0.7)
axis(side=2,cex.axis=0.7,las=2)
} else {
plot(geom,add=T,border='red')
}
}
The figure generated above:
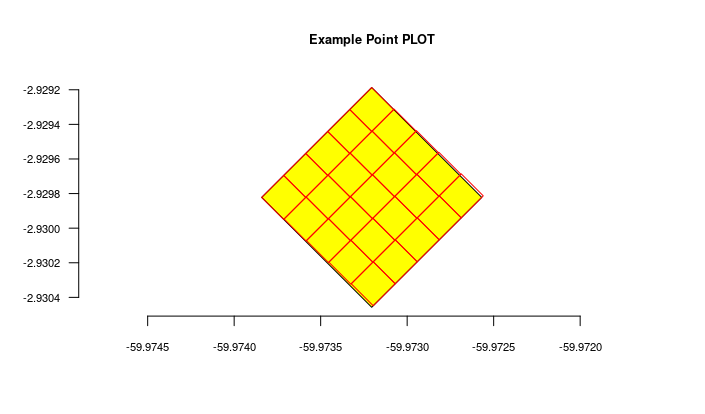
Import transects
This code will import two transects, when defined by a LINESTRING geometry, the other only by a point geometry. See figures below for the imported result
#geometry of transect in Manaus
#read trail from a kml file
#library(rgdal)
#file = "acariquara.kml"
#file.exists(file)
#mykml = readOGR(file)
#library(rgeos)
#geom = rgeos::writeWKT(mykml,byid=TRUE)
#above will output:
geom = "LINESTRING (-59.9616459699999993 -3.0803612500000002, -59.9617394400000023 -3.0805952900000002, -59.9618530300000003 -3.0807376099999999, -59.9621049400000032 -3.0808563200000001, -59.9621949100000009 -3.0809758500000002, -59.9621587999999974 -3.0812666800000001, -59.9621092399999966 -3.0815010400000000, -59.9620656999999966 -3.0816403499999998, -59.9620170600000009 -3.0818584699999998, -59.9620740699999999 -3.0819864099999998)";
#prep data frame
#the y value refer to a buffer in meters applied to the trail
#y is used to validate the insertion of related individuals
to.odb = data.frame(name='A trail-transect example',y=20, notes='OpenDataBio transect example',geom=geom, adm_level = 101,stringsAsFactors=F)
#import
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
odb_import_locations(data=to.odb,odb_cfg=cfg)
#NOW IMPORT A SECOND TRANSECT WITHOUT POINT GEOMETRY
#then you need to inform the x value, which is the transect length
#ODB will map this transect oriented by the azimuth paramater (south in the example below)
#point geometry = start point
geom = "POINT(-59.973841 -2.929822)"
to.odb = data.frame(name='A transect point geometry',x=300, y=20, azimuth=180,notes='OpenDataBio point transect example',geom=geom, adm_level = 101,stringsAsFactors=F)
odb_import_locations(data=to.odb,odb_cfg=cfg)
locais = odb_get_locations(params=list(adm_level=101),odb_cfg = cfg)
locais[,c('id','locationName','parentName','levelName')]
The code above will result in the following two locations:
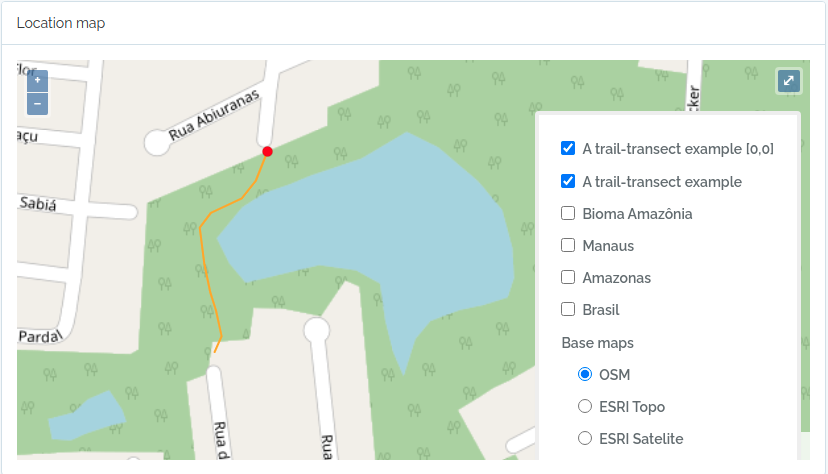
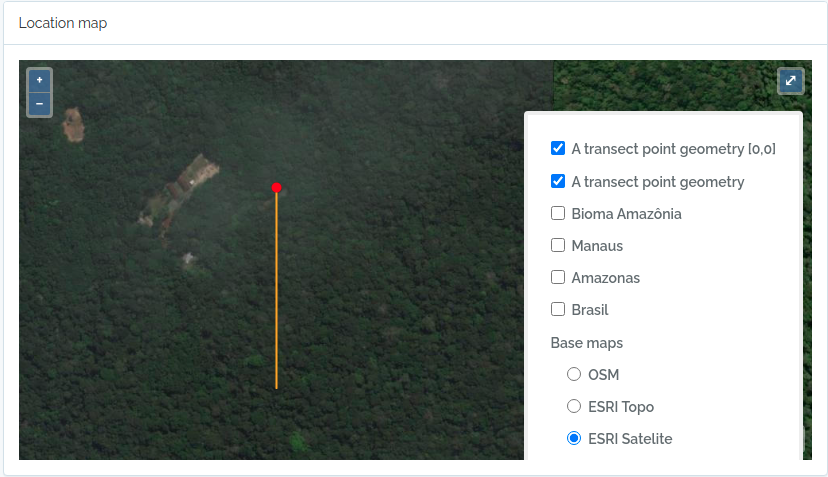
2 - Import BibReferences
Import Bibliography using the OpenDataBio R package
#your connection
library(opendatabio)
base_url="http://localhost/opendatabio/api"
token = “YOUR TOKEN HERE”
cfg = odb_config(base_url=base_url, token = token)
odb_test(cfg)
#read the bibliographic references in R
library(rbibutils)
bibs = readBib(file="yourFileWithReferences.bib")
formatbib <- function(x) {
con <- textConnection("bibref", "w")
writeBib(x,con=con)
bibref = paste(bibref,collapse = " ")
close(con)
return(bibref)
}
#prepare to import to odb
bibtexts = sapply(bibs,formatbib)
data = data.frame(bibtex=bibtexts,standardize=1,stringsAsFactors = F)
#importa
jobid = odb_import_bibreferences(data,odb_cfg = cfg)
#waiting for completion
odb_get_jobs(params=list(id=jobid$id),odb_cfg = cfg)
#get the import log
dt = odb_get_affected_ids(job_id=jobid$id,odb_cfg = cfg)
3 - Import Vernacular
Import Vernacular using the OpenDataBio R package
library(opendatabio)
base_url="http://localhost/opendatabio/api"
token="YOUR TOKEN HERE"
cfg = odb_config(base_url=base_url, token = token)
odb_test(cfg)
#generate fake data for testing
name = c('pau rosa',"casca preciosa")
taxons = c("Aniba roseaodora,Aniba panurensis,Aniba parvifolia","Aniba canelilla")
#get the id of some individuals
inds = odb_get_individuals(params=list(taxon="Aniba roseaodora,Aniba panurensis,Aniba parvifolia",fields='id,scientificName',limit=10),odb_cfg = cfg)
individuals=c(paste(inds$id,collapse = ","),NA)
#idiomas = odb_get_languages(odb_cfg = cfg)
language= c('pt-br','en')
#create a data.frame with this information
verna = data.frame(name,taxons,language,taxons,individuals)
#citations (just generate a data.frame for each vernacular)
umaCitatcao = data.frame(
citation='This would be the cited text',
bibreference="Riberiroetal1999FloraDucke", #bibkey or the id
type='generic', #can be: generic, use, etymology
notes='my observations about this citation')
#add a list-type column
verna$citations = list(oneCitation,NA)
#import to opendatabio
odb_import_vernaculars(verna,odb_cfg = cfg)
4 - Import Media
Import Media using the OpenDataBio R package
Attention
- You basically need the media files in a single folder and a table containing the attributes for each media file;
- See the POST Media API docs to understand which columns can be declared when importing Media. Only 3 columns are mandatory in the attribute table:
filename, object_type, and object_id, but it is important to include additional information about the file, such as date, authors, title, usage license, dataset, and also Tags. - Consider adding at least one keyword (Tag) for each media file.
The method below can be done either through the API using the R package or through the web interface. Test and use whichever is faster to upload the image files.
library(opendatabio)
base_url = "https://opendb.inpa.gov.br/api"
token = "GZ1iXcmRvIFQ"
cfg = odb_config(base_url = base_url)
# path to the folder where the images are stored
folder = 'imagesParaOdb'
# list the file names
filenames = dir(folder, full.names = FALSE)
# read the attribute table
attributes = read.table('arquivoAtributos.csv', sep = ',', header = TRUE, as.is = TRUE, na.strings = c("", "NA", "-"))
# are all files listed in the attribute table?
print(paste(sum(filenames %in% attributes$filename), "of", length(filenames), "files are listed in the attribute table"))
# import to ODB
odb_upload_media_zip(folder = folder, attribute_table = attributes, odb_cfg = cfg)
5 - Import Taxons
Import Taxons using the OpenDataBio R client
Attention
- For a successful importation of published names, you just need to provide a
name attribute, being the fullname for species and infraspecies (e.g. Licaria canella tenuicarpa, Ocotea delicata), same form as GBIF's canonicalName, i.e. do not include infraspecific ranks; - No need to import parent taxons for published names, OpenDataBio does that for you. The import process will use the
name you informed to retrieve taxon info from a nomenclature repository: currently Tropicos, GBIF BackBone Taxonomy and ZOOBANK. A fuzzy search will be performed on the GBIF service if needed. If the taxon name is found, it will automatically get the needed information and also all parent taxa (or accepted names if its tagged as a synonym) as needed and will store in the database for you. If your name is invalid, i.e. it is a synonym of a another name, it will be imported but tagged as invalid. - If you inform
parent and it is different from the one detected by the API the informed parent has preference. - For unpublished species
level, parent, and a person or author_id must also be provided; - The external API will use a fuzzy query while searching GBIF if it does not find an exact match, allowing to detect even when misspelled. However, spelling errors in names may prevent a taxon from being stored.
- Taxonomy is hierarchical and the structure within the OpenDataBio may be treelike. So, you may add, if you want, any node of the tree of life into the Taxon table. For these you need to use the
Clade taxon level. But note, this may not be relevant if you are not going link data to such clade names. - Because the nomenclature check is time consuming, be patient.
OpenDataBio is distributed with a Taxon seed table, that include the root of all kingdoms and some high-level nodes for Plants, i.e. the APWeb Order level tree for Angiosperms.
A simple published name example
The scripts below were tested on top of the OpenDataBio Seed Taxon table, which contains only down to the order level for Angiosperms.
In the taxons table, families Moraceae, Lauraceae and Solanaceae were not yet registered:
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
exists = odb_get_taxons(params=list(root="Moraceae,Lauraceae,Solanaceae"),odb_cfg=cfg)
Returned:
data frame with 0 columns and 0 rows
Now import some species and one infraspecies for the families above, specifying their fullname (canonicalName):
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
spp = c("Ficus schultesii", "Ocotea guianensis","Duckeodendron cestroides","Licaria canella tenuicarpa")
splist = data.frame(name=spp)
odb_import_taxons(splist, odb_cfg=cfg)
Now check with the same code for taxons under those children:
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
exists = odb_get_taxons(params=list(root="Moraceae,Lauraceae,Chrysobalanaceae"),odb_cfg=cfg)
head(exists[,c('id','scientificName', 'taxonRank','taxonomicStatus','parentNameUsage')])
Which will return:
id scientificName taxonRank taxonomicStatus parentName
1 252 Moraceae Family accepted Rosales
2 253 Ficus Genus accepted Moraceae
3 254 Ficus schultesii Species accepted Ficus
4 258 Solanaceae Family accepted Solanales
5 259 Duckeodendron Genus accepted Solanaceae
6 260 Duckeodendron cestroides Species accepted Duckeodendron
7 255 Lauraceae Family accepted Laurales
8 256 Ocotea Genus accepted Lauraceae
9 257 Ocotea guianensis Species accepted Ocotea
10 261 Licaria Genus accepted Lauraceae
11 262 Licaria canella Species accepted Licaria
12 263 Licaria canella subsp. tenuicarpa Subspecies accepted Licaria canella
Note that although we specified only the species and infraspecies names, the API imported also all the needed parent hierarchy up to family, because orders where already registered.
An invalid published name example
The name Licania octandra pallida (Chrysobalanaceae) has been recently turned into a synonym of Leptobalanus octandrus pallidus.
The script below exemplify what happens in such cases.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
#lets check
exists = odb_get_taxons(params=list(root="Chrysobalanaceae"),odb_cfg=cfg)
exists
#in this test returns an empty data frame
#data frame with 0 columns and 0 rows
#now import
spp = c("Licania octandra pallida")
splist = data.frame(name=spp)
odb_import_taxons(splist, odb_cfg=cfg)
#see the results
exists = odb_get_taxons(params=list(root="Chrysobalanaceae"),odb_cfg=cfg)
exists[,c('id','scientificName', 'taxonRank','taxonomicStatus','parentName')]
Which will return:
id scientificName taxonRank taxonomicStatus parentName
1 264 Chrysobalanaceae Family accepted Malpighiales
2 265 Leptobalanus Genus accepted Chrysobalanaceae
3 267 Leptobalanus octandrus Species accepted Leptobalanus
4 269 Leptobalanus octandrus subsp. pallidus Subspecies accepted Leptobalanus octandrus
5 266 Licania Genus accepted Chrysobalanaceae
6 268 Licania octandra Species invalid Licania
7 270 Licania octandra subsp. pallida Subspecies invalid Licania octandra
Note that although we specified only one infraspecies name, the API imported also all the needed parent hierarchy up to family, and because the name is invalid it also imported the acceptedName for this infraspecies and its parents.
An unpublished species or morphotype
It is common to have unpublished local species names (morphotypes) for plants in plots, or yet to be published taxonomic work. Unpublished designation are project specific and therefore MUST also provide an author as different projects may use the same ‘sp.1’ or ‘sp.A’ code for their unpublished taxons.
You may link an unpublished name as any taxon level and do not need to use genus+species logic to assign a morphotype for which the genus or upper level taxonomy is undefined. For example, you may store a ‘species’ level with name ‘Indet sp.1’ and parent_name ‘Laurales’, if the lowest level formal determination you have is the order level. In this example, there is no need to store a Indet genus and Indet family taxons just to account for this unidentified morphotype.
##assign an unpublished name for which you only know belongs to the Angiosperms and you have this node in the Taxon table already
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
#check that angiosperms exist
odb_get_taxons(params=list(name='Angiosperms'),odb_cfg = cfg)
#if it is there, start creating a data.frame to import
to.odb = data.frame(name='Morphotype sp.1', parent='Angiosperms', stringsAsFactors=F)
#get species level numeric code
to.odb$level=odb_taxonLevelCodes('species')
#you must provide an author that is a Person in the Person table. Get from server
odb.persons = odb_get_persons(params=list(search='João Batista da Silva'),odb_cfg=cfg)
#found
head(odb.persons)
#add the author_id to the data.frame
#NOTE it is not author, but author_id or person)
#this makes odb understand it is an unpublished name
to.odb$author_id = odb.persons$id
#import
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
odb_import_taxons(to.odb,odb_cfg = cfg)
Check the imported record:
exists = odb_get_taxons(params=list(name='Morphotype sp.1'),odb_cfg = cfg)
exists[,c('id','scientificName', 'taxonRank','taxonomicStatus','parentName','scientificNameAuthorship')]
Some columns for the imported record:
id scientificName taxonRank taxonomicStatus parentName scientificNameAuthorship
1 276 Morphotype sp.1 Species unpublished Angiosperms João Batista da Silva - Silva, J.B.D.
Import a published clade
You may add a clade Taxon and may reference its publication using the bibkey entry. So, it is possible to actually store all relevant nodes of any phylogeny in the Taxon hierarchy.
#parent must be stored already
odb_get_taxons(params=list(name='Pagamea'),odb_cfg = cfg)
#define clade Taxon
to.odb = data.frame(name='Guianensis core', parent_name='Pagamea', stringsAsFactors=F)
to.odb$level = odb_taxonLevelCodes('clade')
#add a reference to the publication where it is published
#import bib reference to database beforehand
odb_get_bibreferences(params(bibkey='prataetal2018'),odb_cfg=cfg)
to.odb$bibkey = 'prataetal2018'
#then add valid species names as children of this clade instead of the genus level
children = data.frame(name = c('Pagamea guianensis','Pagamea angustifolia','Pagamea puberula'),stringsAsFactors=F)
children$parent_name = 'Guianensis core'
children$level = odb_taxonLevelCodes('species')
children$bibkey = NA
#merge
to.odb = rbind(to.odb,children)
#import
odb_import_taxons(to.odb,odb_cfg = cfg)
6 - Import Persons
Import Persons using the OpenDataBio R client
Check the POST Persons API docs in order to understand which columns can be declared when importing Persons.
It is recommended you use the web interface, as it will warn you in case the person you want to register has a similar, likely the same, person already registered. The API will only check for identical Abbreviations, which is the single restriction of the Person class. Abbreviations are unique and duplications are not allowed. This does not prevent data downloaded from repositories to have different abbreviations or full name for the same person. So, you should standardize secondary data before importing into the server to minimize such common errors.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ"
cfg = odb_config(base_url=base_url, token = token)
one = data.frame(full_name='Adolpho Ducke',abbreviation='DUCKE, A.',notes='Grande botânico da Amazônia',stringsAsFactors = F)
two = data.frame(full_name='Michael John Gilbert Hopkins',abbreviation='HOPKINKS, M.J.G.',notes='Curador herbário INPA',stringsAsFactors = F)
to.odb= rbind(one,two)
odb_import_persons(to.odb,odb_cfg=cfg)
#may also add an email entry if you have one
Get the data
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
cfg = odb_config(base_url=base_url)
persons = odb_get_persons(odb_cfg=cfg)
persons = persons[order(persons$id,decreasing = T),]
head(persons,2)
Will output:
id full_name abbreviation email institution notes
613 1582 Michael John Gilbert Hopkins HOPKINKS, M.J.G. <NA> NA Curador herbário INPA
373 1581 Adolpho Ducke DUCKE, A. <NA> NA Grande botânico da Amazônia
7 - Import Traits
Import Traits using the OpenDataBio R client
Attention
- Strongly recommended you use the web interface to enter traits one by one. Use batch imports only if you have too many.
- Before importing any trait, make sure the variable is not already registered in the system with a different name. Preventing duplication is important as the Trait class is shared among all users of an OpenDataBio installation. This also means that once a trait is used for Measurements, you may only alter the trait’s definition, for example, a category name, if you were the only user entering measurements for the trait. Otherwise, somebody else used that definition and you will not be able to alter it.
- Export_name must be unique in a single ODB installation. This name is used when exporting data and on forms, and you should consider making it as short and informative (of definition) as possible. Also, API will validate the export name - must be camelCase, PascalCase or snake_case, and you cannot add spaces or any special character (accents). Ex. ‘dbh’,‘dbhPom’, ’leafLength’, ‘LeafLength’ or ’leaf_length’, respectively.
Traits can be imported using odb_import_traits().
Read carefully the Traits POST API.
Traits types
See odb_traitTypeCodes() for possible trait types.
Trait name and categories User translations
Fields name and description could be one of following:
- using the Language code as keys:
list("en" = "Diameter at Breast Height","pt-br" ="Diâmetro a Altura do Peito") - or using the Language names as keys:
list("English" ="Diameter at Breast Height","Portuguese" ="Diâmetro a Altura do Peito").
Field categories must include for each category+rank+lang the following fields: lang=mixed - required, the id, code or name of the language of the translationname=string - required, the translated category name required (name+rank+lang must be unique)rank=number - required, rank is important to indicate the same category across languages, and defines ordinal traits;description=string - optional for categories, a definition of the category.
This may be formatted as a data.frame and placed in the categories column of another data.frame:
data.frame(
rbind(
c("lang"="en","rank"=1,"name"="small","description"="smaller than 1 cm"),
c("lang"="pt-br","rank"=1,"name"="pequeno","description"="menor que 1 cm"),
c("lang"="en","rank"=2,"name"="big","description"="bigger than 10 cm"),
c("lang"="pt-br","rank"=2,"name"="grande","description"="maior que 10 cm")
),
stringsAsFactors=FALSE
)
Quantitative trait example
For quantitative traits for either integers or real values (types 0 or 1).
odb_traitTypeCodes()
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#do this first to build a correct data.frame as it will include translations list
to.odb = data.frame(type=1,export_name = "dbh", unit='centimeters',stringsAsFactors = F)
#add translations (note double list)
#format is language_id = translation (and the column be a list with the translation lists)
to.odb$name[[1]]= list('1' = 'Diameter at breast height', '2' = 'Diâmetro à altura do peito')
to.odb$description[[1]]= list('1' = 'Stem diameter measured at 1.3m height','2' = 'Diâmetro do tronco medido à 1.3m de altura')
#measurement validations
to.odb$range_min = 10 #this will restrict the minimum measurement value allowed in the trait
to.odb$range_max = 400 #this will restrict the maximum value
#measurements can be linked to (classes concatenated by , or a list)
to.odb$objects = "Individual,Voucher,Taxon" #makes no sense link such measurements to Locations
to.odb$notes = 'this is quantitative trait example'
#import
odb_import_traits(to.odb,odb_cfg=cfg)
Categorical trait example
- Must include categories. The only difference between ordinal and categorical traits is that ordinal categories will have a rank and the rank will be inferred from the order the categories are informed during importation. Note that ordinal traits are semi-quantitative and so, if you have categories ask yourself whether they are not really ordinal and register accordingly.
- Like the Trait name and description, categories may also have different language translations, and you SHOULD enter the translations for the languages available (
odb_get_languages()) in the server, so the Trait will be accessible in all languages. English is mandatory, so at least the English name must be informed. Categories may have a description associated, but sometimes the category name is self explanatory, so descriptions of categories are not mandatory.
odb_traitTypeCodes()
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#do this first to build a correct data.frame as it will include translations list
#do this first to build a correct data.frame as it will include translations list
to.odb = data.frame(type=3,export_name = "specimenFertility", stringsAsFactors = F)
#trait name and description
to.odb$name = data.frame("en"="Specimen Fertility","pt-br"="Fertilidade do especímene",stringsAsFactors=F)
to.odb$description = data.frame("en"="Kind of reproductive stage of a collected plant","pt-br"="Estágio reprodutivo de uma amostra de planta coletada",stringsAsFactors=F)
#categories (if your trait is ORDINAL, the add categories in the wanted order here)
categories = data.frame(
rbind(
c('en',1,"Sterile"),
c('pt-br',1,"Estéril"),
c('en',2,"Flowers"),
c('pt-br',2,"Flores"),
c('en',3,"Fruits"),
c('pt-br',3,"Frutos"),
c('en',4,"Flower buds"),
c('pt-br',4,"Botões florais")
),
stringsAsFactors =FALSE
)
colnames(categories) = c("lang","rank","name")
#descriptions not included for categories as they are obvious,
# but you may add a 'description' column to the categories data.frame
#objects for which the trait may be used for
to.odb$objects = "Individual,Voucher"
to.odb$notes = 'a fake note for a multiselection categorical trait'
to.odb$categories = list(categories)
#import
odb_import_traits(to.odb,odb_cfg=cfg)
Link type trait example
Link types are traits that allow you link a Taxon or Voucher as a value measurement to another object. For example, you may conduct a plant inventory for which you have only counts for Taxon associated to a locality. Therefore, you may create a LINK trait, which will allow you to store the count values for any Taxon as measurements for a particular location (POINT, POLYGON). Or you may link such values to Vouchers instead of Taxons if you have a representative specimen for the taxons.
Use the WebInterface.
Text and color traits
Text and color traits require the minimum fields only for trait registration. Text traits allow the storage of textual observations. Color will only allow color codes (see example in Importing Measurements)
odb_traitTypeCodes()
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
to.odb = data.frame(type=5,export_name = "taxonDescription", stringsAsFactors = F)
#trait name and description
to.odb$name = data.frame("en"="Taxonomic descriptions","pt-br"="Descrições taxonômicas",stringsAsFactors=F)
to.odb$description = data.frame("en"="Taxonomic descriptions from the literature","pt-br"="Descrições taxonômicas da literatura",stringsAsFactors=F)
#will only be able to use this trait for a measurment associated with a Taxon
to.odb$objects = "Taxon"
#import
odb_import_traits(to.odb,odb_cfg=cfg)
Spectral traits
Spectral traits are specific to spectral data. You must specify the range of wavenumber values for which you may have absorbance or reflectance data, and the length of the spectra to be stored as measurements to allow validation during input. So, for each range and spacement of the spectral values you have, a different SPECTRAL trait must be created.
Use the WebInterface.
8 - Import Individuals & Vouchers
Import Individuals & Vouchers using the OpenDataBio R client
Attention
- You may import vouchers with the Individual POST endpoint. Use the voucher endpoint only for already registered individuals, else follow below to import individuals and their vouchers at once.
- Individuals may have a self taxonomic identification or a non-self taxonomic identification
- Individuals may have multiple locations, but when registering the individual a single location is needed. You may then import additional locations using the Individual-location POST API.
Individuals can be imported using odb_import_individuals() and vouchers with the odb_import_vouchers().
Read carefully the Individual POST API and the Voucher POST API.
Individual example
Prep data for a single individual basic example, representing a tree in a forest plot location.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#the number in the aluminium tag in the forest
to.odb = data.frame(tag='3405.L1', stringsAsFactors=F)
#the collectors (get ids from the server)
(joao = odb_get_persons(params=list(search='joao batista da silva'),odb_cfg=cfg)$id)
(ana = odb_get_persons(params=list(search='ana cristina sega'),odb_cfg=cfg)$id)
#ids concatenated by | pipe
to.odb$collector = paste(joao,ana,sep='|')
#tagged date (lets use an incomplete).
to.odb$date = '2018-07-NA'
#lets place in a Plot location imported with the Location post tutorial
plots = odb_get_locations(params=list(name='A 1ha example plot'),odb_cfg=cfg)
head(plots)
to.odb$location = plots$id
#relative position within parent plot
to.odb$x = 10.4
to.odb$y = 32.5
#or could be
#to.odb$relative_position = paste(x,y,sep=',')
#taxonomic identification
taxon = 'Ocotea guianensis'
#check that exists
(odb_get_taxons(params=list(name='Ocotea guianensis'),odb_cfg=cfg)$id)
#person that identified the individual
to.odb$identifier = odb_get_persons(params=list(search='paulo apostolo'),odb_cfg=cfg)$id
#or you also do to.odb$identifier = "Assunção, P.A.C.L."
#the used form only guarantees the persons is there.
#may add modifers as well [may need to use numeric code instead]
to.odb$modifier = 'cf.'
#or check with to see you spelling is correct
odb_detModifiers()
#and submit the numeric code instaed
to.odb$modifier = 3
#an incomplete identification date
to.odb$identification_date = list(year=2005)
#or to.odb$identification_date = "2005-NA-NA"
Lets import the above record:
odb_import_individuals(to.odb,odb_cfg = cfg)
#lets import this individual
odb_import_individuals(to.odb,odb_cfg = cfg)
#check the job status
odb_get_jobs(params=list(id=130),odb_cfg = cfg)
Ops, I forgot to inform a dataset and my user does not have a default dataset defined.
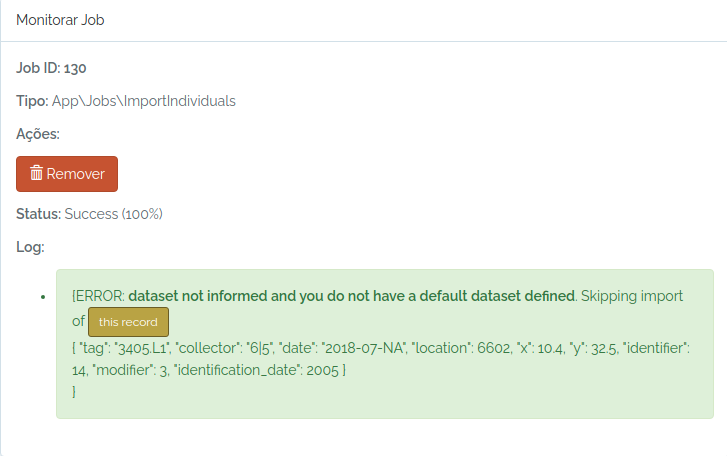
So, I just inform an existing dataset and try again:
dataset = odb_get_datasets(params=list(name="Dataset test"),odb_cfg=cfg)
dataset
to.odb$dataset = dataset$id
odb_import_individuals(to.odb,odb_cfg = cfg)
The individual was imported. The image below shows the individual (yellow dot) mapped in the plot:
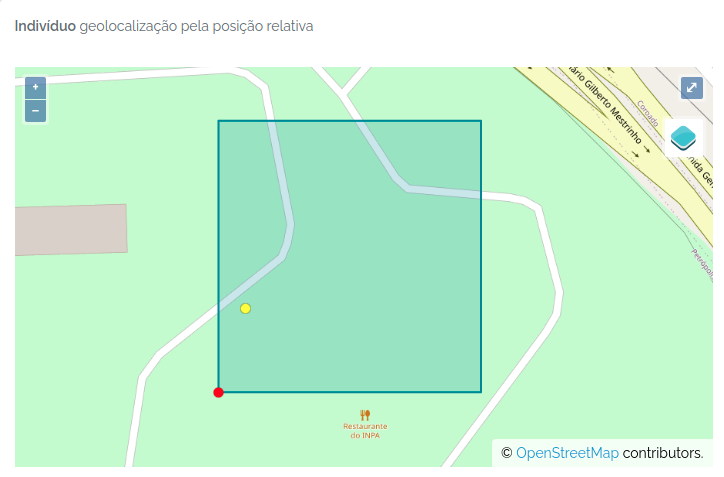
Importing Individuals and Vouchers at once
Individuals are the actual object that has most of the information related to Vouchers, which are samples in a Biocollection. Therefore, you may import an individual record with the specification of one or more vouchers.
#a fake plant record somewhere in the Amazon
aplant = data.frame(taxon="Duckeodendron cestroides", date="2021-09-09", latitude=-2.34, longitude=-59.845,angle=NA,distance=NA, collector="Oliveira, A.A. de|João Batista da Silva", tag="3456-A",dataset=1)
#a fake set of vouchers for this individual
herb = data.frame(biocollection=c("INPA","NY","MO"),biocollection_number=c("12345A","574635","ANOTHER FAKE CODE"),biocollection_type=c(2,3,3))
#add this dataframe to the object
aplant$biocollection = NA
aplant$biocollection = list(herb)
#another fake plant
asecondplant = data.frame(taxon="Ocotea guianensis", date="2021-09-09", latitude=-2.34, longitude=-59.89,angle=240,distance=50, collector="Oliveira, A.A. de|João Batista da Silva", tag="3456",dataset=1)
asecondplant$biocollection = NA
#merge the fake data
to.odb = rbind(aplant,asecondplant)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
odb_import_individuals(to.odb, odb_cfg=cfg)
Check the imported data
The script above has created records for both the Individual and Voucher model:
#get the imported individuals using a wildcard
inds = odb_get_individuals(params = list(tag='3456*'),odb_cfg = cfg)
inds[,c("basisOfRecord","scientificName","organismID","decimalLatitude","decimalLongitude","higherGeography") ]
Will return:
basisOfRecord scientificName organismID decimalLatitude decimalLongitude higherGeography
1 Organism Ocotea guianensis 3456 - Oliveira - UnnamedPoint_5989234 -2.3402 -59.8904 Brasil | Amazonas | Rio Preto da Eva
2 Organism Duckeodendron cestroides 3456-A - Oliveira - UnnamedPoint_5989234 -2.3400 -59.8900 Brasil | Amazonas | Rio Preto da Eva
And the vouchers:
#get the vouchers imported with the first plant data
vouchers = odb_get_vouchers(params = list(individual=inds$id),odb_cfg = cfg)
vouchers[,c("basisOfRecord","scientificName","organismID","collectionCode","catalogNumber") ]
Will return:
basisOfRecord scientificName occurrenceID collectionCode catalogNumber
1 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -INPA.12345A INPA 12345A
2 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -MO.ANOTHER FAKE CODE MO ANOTHER FAKE CODE
3 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -NY.574635 NY 574635
Import Vouchers for Existing Individuals
Attention
- Vouchers are samples of Individuals deposited in BioCollections. If the Biocollection is a genetic resource, the Voucher may just represent a DNA or tissue sample.
- Therefore, Vouchers must belong to an Individual and to a BioCollection, which are the mandatory information to register vouchers. Collectors, collection date, location and taxonomic identity may just be extracted from the Individual’s record.
- A Voucher may have its own collector, collector number and collecting date, different from the Individual it belongs to, when needed.
The mandatory fields are:
individual = individual id or fullname (organismID);biocollection = acronym or id of the BioCollection - use odb_get_biocollections() to check if it is registered, otherwise, first store the BioCollection in in the database;
For additional fields see Voucher POST API.
A simple voucher import
#a holotype voucher with same collector and date as individual
onevoucher = data.frame(individual=1,biocollection="INPA",biocollection_number=1234,biocollection_type=1,dataset=1)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
odb_import_vouchers(onevoucher, odb_cfg=cfg)
#get the imported voucher
voucher = odb_get_vouchers(params=list(individual=1),cfg)
vouchers[,c("basisOfRecord","scientificName","occurrenceID","collectionCode","catalogNumber") ]
Different voucher for an individual
Two vouchers for the same individual, one with the same collector and date as the individual, the other at different time and by other collectors.
#one with same date and collector as individual
one = data.frame(individual=2,biocollection="INPA",biocollection_number=1234,dataset=1,collector=NA,number=NA,date=NA)
#this one with different collector and date
two= data.frame(individual=2,biocollection="INPA",biocollection_number=4435,dataset=1,collector="Oliveira, A.A. de|João Batista da Silva",number=3456,date="1991-08-01")
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
to.odb = rbind(one,two)
odb_import_vouchers(to.odb, odb_cfg=cfg)
#get the imported voucher
voucher = odb_get_vouchers(params=list(individual=2),cfg)
voucher[,c("basisOfRecord","scientificName","occurrenceID","collectionCode","catalogNumber") ]
Output of imported records:
basisOfRecord scientificName occurrenceID collectionCode catalogNumber recordedByMain
1 PreservedSpecimens Unidentified plot tree - Vicentini -INPA.1234 INPA 1234 Vicentini, A.
2 PreservedSpecimens Unidentified 3456 - Oliveira -INPA.4435 INPA 4435 Oliveira, A.A. de
9 - Import Measurements
Import Measurements using the OpenDataBio R client
Attention
- Measurement have one restriction: duplicated measurement values or categories for the same object_id and same date will not be imported unless you specify a
duplicated=1 with for the record. - Measurements are straightforward, but you need to grap some info from other models to import them. You need to provide a
dataset for which you do have permissions, trait_id (export_name or id), value (content varies depending on trait type), date (a complete or incomplete date), person (person that measured), object_type (one of taxon, individual, voucher or location) and object_id (its odb id). For LINK trait type you must provide a link_id column, and ‘value’ becomes optional for this specific case. - TIP: To link among measurements for different traits you may add standard keys to the
note measurement field. For example, you measure LeafLength and LeafWidth for the same leaves, so in both measurements add “leaf1” ,“leaf2” to the note field to link them. Also, if you measure DBH and DBH.POM in a forest plot inventory and have several stems for a plant, you may add “stem1”,“stem2” to permit matching a stem DBH to its specific height of measurement. This kind of link may be useful in analyses. - Several validations will happen during measurement importation. Consider verifying the data locally before submitting a job to facilitate troubleshooting, and guarantee the values are consistent with your expectations. You will not be able to UPDATE nor DELETE measurements with the API, so, avoid importing several measurements that will require fixing afterwards (one by one).
- For Quantitative Traits, OpenDataBio cannot validate that your measurements are in the same unit as the Trait definition. Therefore, check which unit the trait has, and convert your data accordingly before uploading.
Measurements can be imported using odb_import_measurements(). Read carefully the Measurements POST API.
Quantitative measurements
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#get the trait id from the server (check that trait exists)
#generate some fake data for 10 measurements
dbhs = sample(seq(10,100,by=0.1),10)
object_ids = sample(1:3,length(dbhs),replace=T)
dates = sample(as.Date("2000-01-01"):as.Date("2000-03-31"),length(dbhs))
dates = lapply(dates,as.Date,origin="1970-01-01")
dates = lapply(dates,as.character)
dates = unlist(dates)
to.odb = data.frame(
trait_id = 'dbh',
value = dbhs,
date = dates,
object_type = 'Individual',
object_id=object_ids,
person="Oliveira, A.A. de",
dataset = 1,
notes = "some fake measurements",
stringsAsFactors=F)
#this will only work if the person exists, the individual ids exist
#and if the trait with export_name=dbh exist
odb_import_measurements(to.odb,odb_cfg=cfg)
Get the imported data:
dad = odb_get_measurements(params = list(dataset=1),odb_cfg=cfg)
dad[,c("id","basisOfRecord", "measured_type", "measured_id", "measurementType",
"measurementValue", "measurementUnit", "measurementDeterminedDate",
"datasetName", "license")]
id basisOfRecord measured_type measured_id measurementType measurementValue measurementUnit measurementDeterminedDate
1 1 MeasurementsOrFact App\\Models\\Individual 3 dbh 86.8 centimeters 2000-02-19
2 2 MeasurementsOrFact App\\Models\\Individual 2 dbh 84.8 centimeters 2000-03-25
3 3 MeasurementsOrFact App\\Models\\Individual 2 dbh 65.7 centimeters 2000-03-15
4 4 MeasurementsOrFact App\\Models\\Individual 3 dbh 88.0 centimeters 2000-03-05
5 5 MeasurementsOrFact App\\Models\\Individual 3 dbh 35.3 centimeters 2000-01-04
6 6 MeasurementsOrFact App\\Models\\Individual 2 dbh 36.0 centimeters 2000-03-23
7 7 MeasurementsOrFact App\\Models\\Individual 2 dbh 78.6 centimeters 2000-03-22
8 8 MeasurementsOrFact App\\Models\\Individual 2 dbh 69.7 centimeters 2000-03-09
9 9 MeasurementsOrFact App\\Models\\Individual 3 dbh 12.3 centimeters 2000-01-30
10 10 MeasurementsOrFact App\\Models\\Individual 3 dbh 14.7 centimeters 2000-01-18
datasetName license
1 Dataset test CC-BY 4.0
2 Dataset test CC-BY 4.0
3 Dataset test CC-BY 4.0
4 Dataset test CC-BY 4.0
5 Dataset test CC-BY 4.0
6 Dataset test CC-BY 4.0
7 Dataset test CC-BY 4.0
8 Dataset test CC-BY 4.0
9 Dataset test CC-BY 4.0
10 Dataset test CC-BY 4.0
Categorical measurements
Categories MUST be informed by their ids or name in the value field. For CATEGORICAL or ORDINAL traits, value must be single value. For CATEGORICAL_MULTIPLE, value may be one or multiple categories ids or names separated by one of | or ; or ,.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#a categorical trait
(odbtraits = odb_get_traits(params=list(name="specimenFertility"),odb_cfg = cfg))
#base line
to.odb = data.frame(trait_id = odbtraits$id, date = '2021-07-31', stringsAsFactors=F)
#the plant was collected with both flowers and fruits, so the value are the two categories
value = c("Flowers","Fruits")
#get categories for this trait if found
(cats = odbtraits$categories[[1]])
#check that your categories are registered for the trait and get their ids
value = cats[match(value,cats$name),'id']
#make multiple categories ids a string value
value = paste(value,collapse=",")
to.odb$value = value
#this links to a voucher
to.odb$object_type = "Voucher"
#get voucher id from API (must be ID).
#Search for collection number 1234
odbspecs = odb_get_vouchers(params=list(number="3456-A"),odb_cfg=cfg)
to.odb$object_id = odbspecs$id[1]
#get dataset id
odbdatasets = odb_get_datasets(params=list(name='Dataset test'),odb_cfg=cfg)
head(odbdatasets)
to.odb$dataset = odbdatasets$id
#person that measured
odbperson = odb_get_persons(params=list(search='ana cristina sega'),odb_cfg=cfg)
to.odb$person = odbperson$id
#import'
odb_import_measurements(to.odb,odb_cfg=cfg)
#get imported
dad = odb_get_measurements(params = list(voucher=odbspecs$id[1]),odb_cfg=cfg)
dad[,c("id","basisOfRecord", "measured_type", "measured_id", "measurementType",
"measurementValue", "measurementUnit", "measurementDeterminedDate",
"datasetName", "license")]
id basisOfRecord measured_type measured_id measurementType measurementValue measurementUnit
1 11 MeasurementsOrFact App\\Models\\Voucher 1 specimenFertility Flowers, Fruits NA
measurementDeterminedDate datasetName license
1 2021-07-31 Dataset test CC-BY 4.0
Color measurements
For color values you have to enter color as their hex RGB strings codes, so they can be rendered graphically and in the web interface. Therefore, any color value is allowed, and it would be easier to use the palette colors in the web interface to enter such measurements. Package gplots allows you to convert color names to hex RGB codes if you want to do it through the API.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#get the trait id from the server (check that trait exists)
odbtraits = odb_get_traits(odb_cfg=cfg)
(m = match(c("fruitColor"),odbtraits$export_name))
#base line
to.odb = data.frame(trait_id = odbtraits$id[m], date = '2014-01-13', stringsAsFactors=F)
#get color value
#install.packages("gplots",dependencies = T)
library(gplots)
(value = col2hex("red"))
to.odb$value = value
#this links to a specimen
to.odb$object_type = "Individual"
#get voucher id from API (must be ID). Search for collection number 1234
odbind = odb_get_individuals(params=list(tag='3456'),odb_cfg=cfg)
odbind$scientificName
to.odb$object_id = odbind$id[1]
#get dataset id
odbdatasets = odb_get_datasets(params=list(name='Dataset test'),odb_cfg=cfg)
head(odbdatasets)
to.odb$dataset = odbdatasets$id
#person that measured
odbperson = odb_get_persons(params=list(search='ana cristina sega'),odb_cfg=cfg)
to.odb$person = odbperson$id
odb_import_measurements(to.odb,odb_cfg=cfg)

Database link type measurements
The LINK trait type allows one to register count data, as for example the number of individuals of a species at a particular location. You have to provide the linked object (link_id), which may be a Taxon or a Voucher depending on the trait definition, and then value recieves the numeric count.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#get the trait id from the server (check that trait exists)
odbtraits = odb_get_traits(odb_cfg=cfg)
(m = match(c("taxonCount"),odbtraits$export_name))
#base line
to.odb = data.frame(trait_id = odbtraits$id[m], date = '2014-01-13', stringsAsFactors=F)
#the taxon to link the count value
odbtax = odb_get_taxons(params=list(name='Ocotea guianensis'),odb_cfg=cfg)
to.odb$link_id = odbtax$id
#now add the count value for this trait type
#this is optional for this measurement,
#however, it would make no sense to include such link without a count in this example
to.odb$value = 23
#a note to clarify the measurement (optional)
to.odb$notes = 'No voucher, field identification'
#this measurement will link to a location
to.odb$object_type = "Location"
#get location id from API (must be ID).
#lets add this to a transect
odblocs = odb_get_locations(params=list(adm_level=101,limit=1),odb_cfg=cfg)
to.odb$object_id = odblocs$id
#get dataset id
odbdatasets = odb_get_datasets(params=list(name='Dataset test'),odb_cfg=cfg)
head(odbdatasets)
to.odb$dataset = odbdatasets$id
#person that measured
odbperson = odb_get_persons(params=list(search='ana cristina sega'),odb_cfg=cfg)
to.odb$person = odbperson$id
odb_import_measurements(to.odb,odb_cfg=cfg)

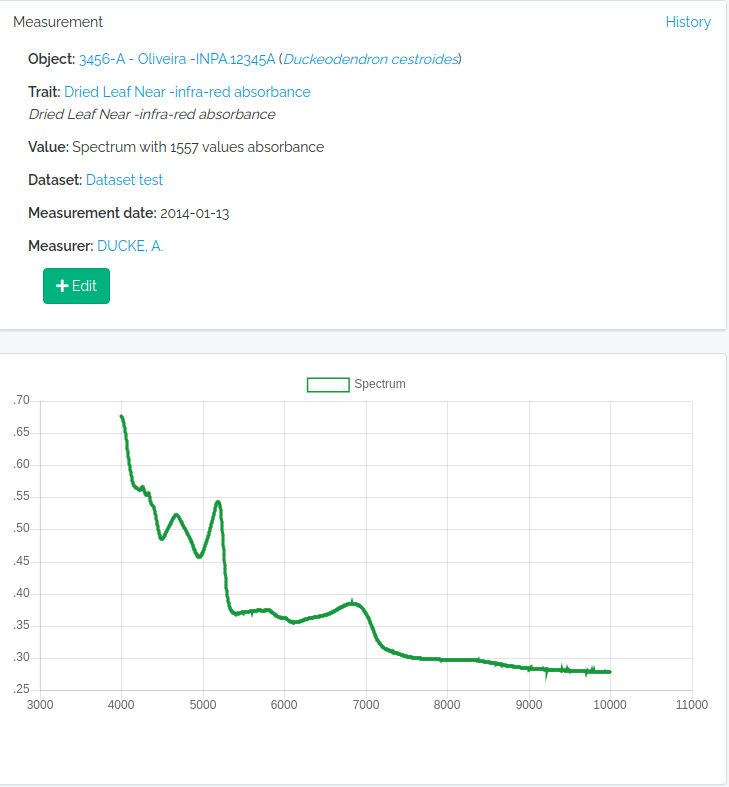
Spectral measurements
value must be a string of spectrum values separated by “;”. The number of concatenated values must match the Trait value_length attribute of the trait, which is extracted from the wavenumber range specification for the trait. So, you may easily check this before importing with odb_get_traits(params=list(fields='all',type=9),cfg)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#read a spectrum
spectrum = read.table("1_Sample_Planta-216736_TAG-924-1103-1_folha-1_abaxial_1.csv",sep=",")
#second column are NIR leaf absorbance values
#the spectrum has 1557 values
nrow(spectrum)
#[1] 1557
#collapse to single string
value = paste(spectrum[,2],collapse = ";")
substr(value,1,100)
#[1] "0.6768057;0.6763237;0.6755353;0.6746023;0.6733549;0.6718447;0.6701176;0.6682984;0.6662288;0.6636459;"
#get the trait id from the server (check that trait exists)
odbtraits = odb_get_traits(odb_cfg=cfg)
(m = match(c("driedLeafNirAbsorbance"),odbtraits$export_name))
#see the trait
odbtraits[m,c("export_name", "unit", "range_min", "range_max", "value_length")]
#export_name unit range_min range_max value_length
#6 driedLeafNirAbsorbance absorbance 3999.64 10001.03 1557
#must be true
odbtraits$value_length[m]==nrow(spectrum)
#[1] TRUE
#base line
to.odb = data.frame(trait_id = odbtraits$id[m], value=value, date = '2014-01-13', stringsAsFactors=F)
#this links to a voucher
to.odb$object_type = "Voucher"
#get voucher id from API (must be ID).
#search for a collection number
odbspecs = odb_get_vouchers(params=list(number="3456-A"),odb_cfg=cfg)
to.odb$object_id = odbspecs$id[1]
#get dataset id
odbdatasets = odb_get_datasets(params=list(name='Dataset test'),odb_cfg=cfg)
to.odb$dataset = odbdatasets$id
#person that measured
odbperson = odb_get_persons(params=list(search='adolpho ducke'),odb_cfg=cfg)
to.odb$person = odbperson$id
#import
odb_import_measurements(to.odb,odb_cfg=cfg)
Text measurements
Just add the text to the value field and proceed as for the other trait types.










