Import Individuals & Vouchers
5 minute read
Individuals can be imported using odb_import_individuals() and vouchers with the odb_import_vouchers().
Read carefully the Individual POST API and the Voucher POST API.
Individual example
Prep data for a single individual basic example, representing a tree in a forest plot location.
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
#the number in the aluminium tag in the forest
to.odb = data.frame(tag='3405.L1', stringsAsFactors=F)
#the collectors (get ids from the server)
(joao = odb_get_persons(params=list(search='joao batista da silva'),odb_cfg=cfg)$id)
(ana = odb_get_persons(params=list(search='ana cristina sega'),odb_cfg=cfg)$id)
#ids concatenated by | pipe
to.odb$collector = paste(joao,ana,sep='|')
#tagged date (lets use an incomplete).
to.odb$date = '2018-07-NA'
#lets place in a Plot location imported with the Location post tutorial
plots = odb_get_locations(params=list(name='A 1ha example plot'),odb_cfg=cfg)
head(plots)
to.odb$location = plots$id
#relative position within parent plot
to.odb$x = 10.4
to.odb$y = 32.5
#or could be
#to.odb$relative_position = paste(x,y,sep=',')
#taxonomic identification
taxon = 'Ocotea guianensis'
#check that exists
(odb_get_taxons(params=list(name='Ocotea guianensis'),odb_cfg=cfg)$id)
#person that identified the individual
to.odb$identifier = odb_get_persons(params=list(search='paulo apostolo'),odb_cfg=cfg)$id
#or you also do to.odb$identifier = "Assunção, P.A.C.L."
#the used form only guarantees the persons is there.
#may add modifers as well [may need to use numeric code instead]
to.odb$modifier = 'cf.'
#or check with to see you spelling is correct
odb_detModifiers()
#and submit the numeric code instaed
to.odb$modifier = 3
#an incomplete identification date
to.odb$identification_date = list(year=2005)
#or to.odb$identification_date = "2005-NA-NA"
Lets import the above record:
odb_import_individuals(to.odb,odb_cfg = cfg)
#lets import this individual
odb_import_individuals(to.odb,odb_cfg = cfg)
#check the job status
odb_get_jobs(params=list(id=130),odb_cfg = cfg)
Ops, I forgot to inform a dataset and my user does not have a default dataset defined.
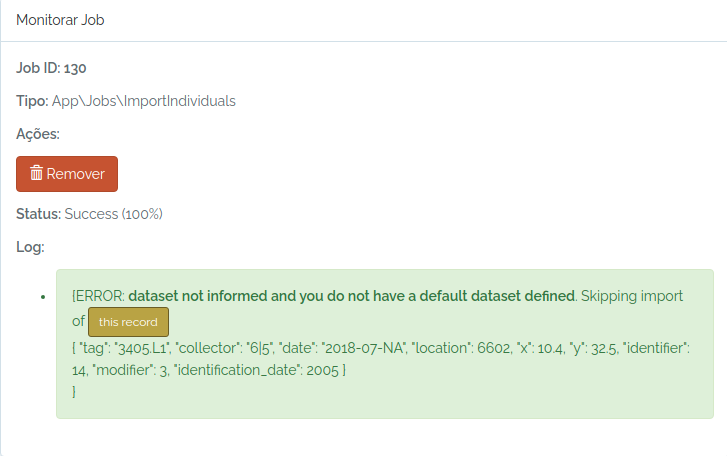
So, I just inform an existing dataset and try again:
dataset = odb_get_datasets(params=list(name="Dataset test"),odb_cfg=cfg)
dataset
to.odb$dataset = dataset$id
odb_import_individuals(to.odb,odb_cfg = cfg)
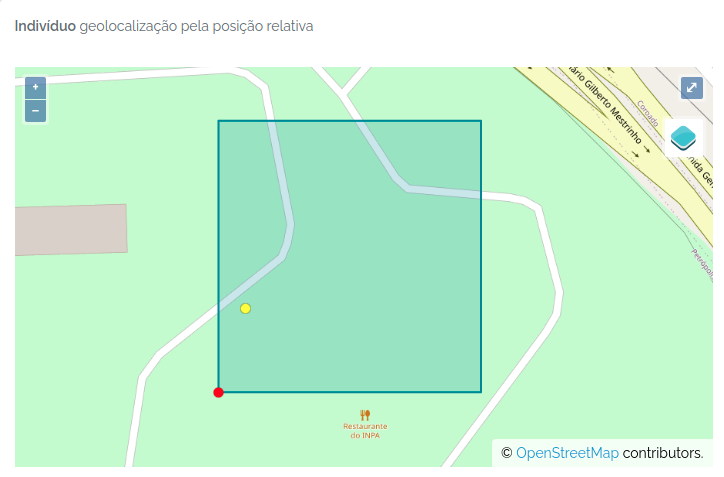
The individual was imported. The image below shows the individual (yellow dot) mapped in the plot:
Importing Individuals and Vouchers at once
Individuals are the actual object that has most of the information related to Vouchers, which are samples in a Biocollection. Therefore, you may import an individual record with the specification of one or more vouchers.
#a fake plant record somewhere in the Amazon
aplant = data.frame(taxon="Duckeodendron cestroides", date="2021-09-09", latitude=-2.34, longitude=-59.845,angle=NA,distance=NA, collector="Oliveira, A.A. de|João Batista da Silva", tag="3456-A",dataset=1)
#a fake set of vouchers for this individual
herb = data.frame(biocollection=c("INPA","NY","MO"),biocollection_number=c("12345A","574635","ANOTHER FAKE CODE"),biocollection_type=c(2,3,3))
#add this dataframe to the object
aplant$biocollection = NA
aplant$biocollection = list(herb)
#another fake plant
asecondplant = data.frame(taxon="Ocotea guianensis", date="2021-09-09", latitude=-2.34, longitude=-59.89,angle=240,distance=50, collector="Oliveira, A.A. de|João Batista da Silva", tag="3456",dataset=1)
asecondplant$biocollection = NA
#merge the fake data
to.odb = rbind(aplant,asecondplant)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
odb_import_individuals(to.odb, odb_cfg=cfg)
Check the imported data
The script above has created records for both the Individual and Voucher model:
#get the imported individuals using a wildcard
inds = odb_get_individuals(params = list(tag='3456*'),odb_cfg = cfg)
inds[,c("basisOfRecord","scientificName","organismID","decimalLatitude","decimalLongitude","higherGeography") ]
Will return:
basisOfRecord scientificName organismID decimalLatitude decimalLongitude higherGeography
1 Organism Ocotea guianensis 3456 - Oliveira - UnnamedPoint_5989234 -2.3402 -59.8904 Brasil | Amazonas | Rio Preto da Eva
2 Organism Duckeodendron cestroides 3456-A - Oliveira - UnnamedPoint_5989234 -2.3400 -59.8900 Brasil | Amazonas | Rio Preto da Eva
And the vouchers:
#get the vouchers imported with the first plant data
vouchers = odb_get_vouchers(params = list(individual=inds$id),odb_cfg = cfg)
vouchers[,c("basisOfRecord","scientificName","organismID","collectionCode","catalogNumber") ]
Will return:
basisOfRecord scientificName occurrenceID collectionCode catalogNumber
1 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -INPA.12345A INPA 12345A
2 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -MO.ANOTHER FAKE CODE MO ANOTHER FAKE CODE
3 PreservedSpecimens Duckeodendron cestroides 3456-A - Oliveira -NY.574635 NY 574635
Import Vouchers for Existing Individuals
The mandatory fields are:
individual= individual id or fullname (organismID);biocollection= acronym or id of the BioCollection - useodb_get_biocollections()to check if it is registered, otherwise, first store the BioCollection in in the database;
For additional fields see Voucher POST API.
A simple voucher import
#a holotype voucher with same collector and date as individual
onevoucher = data.frame(individual=1,biocollection="INPA",biocollection_number=1234,biocollection_type=1,dataset=1)
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
odb_import_vouchers(onevoucher, odb_cfg=cfg)
#get the imported voucher
voucher = odb_get_vouchers(params=list(individual=1),cfg)
vouchers[,c("basisOfRecord","scientificName","occurrenceID","collectionCode","catalogNumber") ]
Different voucher for an individual
Two vouchers for the same individual, one with the same collector and date as the individual, the other at different time and by other collectors.
#one with same date and collector as individual
one = data.frame(individual=2,biocollection="INPA",biocollection_number=1234,dataset=1,collector=NA,number=NA,date=NA)
#this one with different collector and date
two= data.frame(individual=2,biocollection="INPA",biocollection_number=4435,dataset=1,collector="Oliveira, A.A. de|João Batista da Silva",number=3456,date="1991-08-01")
library(opendatabio)
base_url="https://opendb.inpa.gov.br/api"
token ="GZ1iXcmRvIFQ" #this must be your token not this value
cfg = odb_config(base_url=base_url, token = token)
to.odb = rbind(one,two)
odb_import_vouchers(to.odb, odb_cfg=cfg)
#get the imported voucher
voucher = odb_get_vouchers(params=list(individual=2),cfg)
voucher[,c("basisOfRecord","scientificName","occurrenceID","collectionCode","catalogNumber") ]
Output of imported records:
basisOfRecord scientificName occurrenceID collectionCode catalogNumber recordedByMain
1 PreservedSpecimens Unidentified plot tree - Vicentini -INPA.1234 INPA 1234 Vicentini, A.
2 PreservedSpecimens Unidentified 3456 - Oliveira -INPA.4435 INPA 4435 Oliveira, A.A. de